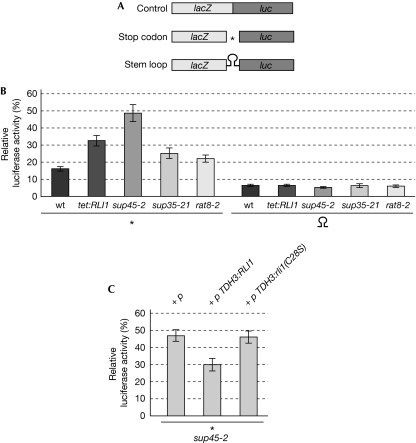
Figure 4.
Rli1 is required for efficient stop codon recognition. (A) Scheme of the reporter constructs used to determine the read-through activity. (B) Read-through activities are shown for wild type, a strain that downregulates the RLI1 expression (tet:RLI1), sup45-2, sup35-21 and rat8-2, encoding the defective DEAD-box RNA helicase Dbp5. All strains carrying either of the reporter constructs were grown to log phase and incubated at 37°C for 15 min before cell lysis. β-Galactosidase and luciferase activities were measured and their ratios were used to calculate the relative molar luciferase expression. (C) Suppression of the termination read-through defects of sup45-2 by increased Rli1 level (TDH3:RLI1), but not an Fe–S cluster mutant (TDH3:rli1(C28S)), is shown. All strains carrying the reporter construct and the indicated plasmids were treated as described in (B). The results of at least 10 independent experiments are shown. The asterisks represent the termination codon UAG. Rli1, RNase L inhibitor; wt, wild type.