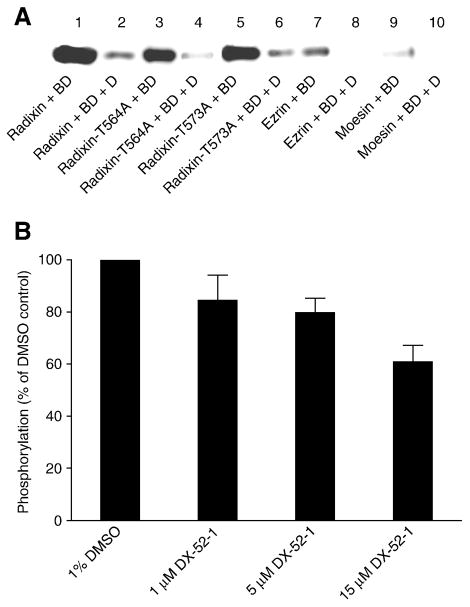
Figure 8. Thr 564 and Thr 573 are not the sites of modification by DX-52-1 on radixin, and ezrin and moesin are also modified by DX-52-1, but much less intensely than radixin.
(A) DX-52-1 binds the Thr-to-Ala radixin mutants (radixin-T564A and radixin-T573A), along with wild-type radixin, but only very weakly binds ezrin and moesin. DX-52-1-binding experiments were performed as described in Materials and Methods with 23 μM of each purified recombinant protein and 10 μM biotinylated DX-52-1 with or without 500 μM of free, unlabeled DX-52-1 (added simultaneously as competitor to verify saturability and specificity). Proteins and treatments are: wild-type radixin, biotinylated DX-52-1 (lane 1); wild-type radixin, biotinylated DX-52-1 (BD) with 50× free, unlabeled DX-52-1 (D; lane 2); radixin-T564A, biotinylated DX-52-1 (lane 3); radixin-T564A, biotinylated DX-52-1 with 50× DX-52-1 (lane 4); radixin-T573A, biotinylated DX-52-1 (lane 5); radixin-T573A, biotinylated DX-52-1 with 50× DX-52-1 (lane 6); ezrin, biotinylated DX-52-1 (lane 7); ezrin, DX-52-1 with 50× DX-52-1 (lane 8); moesin, biotinylated DX-52-1 (lane 9); moesin, biotinylated DX-52-1 with 50× DX-52-1 (lane 10). (B) DX-52-1 does not block phosphorylation of radixin by GRK2. Purified recombinant GRK2 was preincubated with different concentrations of DX-52-1, followed by assay of kinase activity toward radixin, as described in Materials and Methods. Levels (mean and SEM) of [γ-32P]-ATP-labeled radixin were quantitated from autoradiograms from three independent experiments, with values expressed as a percent of the level of the phosphorylated radixin signal for DMSO controls (normalization to the parallel control for each experiment prior to statistics).