Abstract
Objective
Autophagy is a process for turnover of intracellular organelles and molecules that protects cells during stress responses. This study evaluated the potential role of ULK1, an inducer of autophagy, Beclin1, a regulator of autophagy and LC3, which executes autophagy, in the development of osteoarthritis (OA) and in cartilage cell death.
Methods
Expression of ULK1, Beclin1 and LC3 were analyzed in normal and OA human articular cartilage and in knee joints of mice with aging-related and surgically induced OA by using immunohistochemistry (IHC) and western blotting. Poly-ADP(ribose) polymerase (Parp p85) was used to determine the correlation between cell death and autophagy.
Results
In normal human articular cartilage ULK1, Beclin1 and LC3 were constitutively expressed. ULK1, Beclin1 and LC3 protein expression were reduced in OA chondrocytes and cartilage but these three proteins were strongly expressed in the OA cell clusters. In mouse knee joints loss of glycosaminoglycans (GAGs) was observed at 9 and 12 months of age and in the surgical OA model 8 weeks after knee destabilization. Expression of ULK1, Beclin1 and LC3 decreased together with GAG loss while Parp p85 was increased.
Conclusion
Autophagy may be a protective or homeostatic mechanism in normal cartilage. By contrast, human OA, aging-related and surgically-induced OA in mice are associated with a reduction and loss of ULK1, Beclin1 and LC3 expression and a related increase in apoptosis. These results suggest that compromised autophagy represents a novel mechanism in the development of OA.
Keywords: Osteoarthritis, Aging, Autophagy, Cartilage, Cell Death
INTRODUCTION
Disruption of the articular cartilage surface, degradation of extracellular matrix (ECM) and reduced cartilage cellularity, are major histological features of osteoarthritis (OA), the most common aging-related joint pathology (1,2). Because chondrocytes maintain the dynamic equilibrium between production of the ECM and its enzymatic degradation, the molecular mechanisms that control cell fate in the cartilage are important to be uncovered (3). Many factors can be implicated in the development of OA but one of the most important risk factors is aging (4). Aging is a process characterized by a progressive accumulation of damaged macromolecules and organelles in somatic cells during the post-developmental period, leading to the decreased ability of cells to function normally and survive (5). However, mechanisms leading to the aging-related cartilage degeneration remain to be determined.
Macroautophagy is a major physiological mechanism that targets altered and dysfunctional cytosolic macromolecules, membranes and organelles for delivery to lysosomes for degradation and recycling (5,6,7,8). Atg genes control the autophagy process leading to the induction and nucleation of autophagic vesicles, their expansion and fusion with lysosomes, allowing enzymatic degradation and recycling (9,10). Among the Atg genes, Atg1, Atg6 and Atg8 (ULK1, Beclin1 and LC3 in mammals, respectively) are three major regulators of the autophagy pathway. ULK1 is a key intermediate in the transduction of pro-autophagic signals to autophagosome formation (11). Beclin1 forms a complex with type III PI3 kinase and Vps34 that allows nucleation of the autophagic vesicle (12). Finally, the formation and expansion of the autophagosome requires two protein conjugation systems that involve the Atg proteins LC3 and Atg12 (13). LC3 is present in two forms, LC3-1 in the cytoplasm and LC3-II bound to the autophagosome membrane. During autophagy LC3-I is converted to LC3-II through lipidation by a ubiquitin-like system resulting in the association of LC3-II with autophagy vesicles. The amount of LC3-II is correlated with the extent of autophagosome formation (14). Autophagy plays a fundamental role in cellular homeostasis and functions primarily to promote cellular and organism health. In certain physiological and pathological conditions it can also lead to a form of cell death that is characterized by cytoplasmic vacuolation and termed type II programmed cell death or cell death by autophagy (15,16,17). In most experimental models suppression of autophagy genes leads to cell death, indicating a protective and survival-promoting function of autophagy (18,19,20).
In articular cartilage, which is characterized by a very low rate of cell turnover (21) this mechanism would appear to be essential to maintain cellular integrity, function and survival. Furthermore, while autophagy changes in various models and tissues with aging (22), this has not been investigated in articular cartilage. In the present study, we demonstrate that autophagy is a constitutively active and apparently protective process for the maintenance of the homeostatic state in normal cartilage. By contrast, human OA and aging-related and surgically-induced OA in mice are associated with a reduction and loss of ULK1, Beclin1 and LC3 expression in articular cartilage. Furthermore, the reduction of these key regulators of autophagy is accompanied by an increase in apoptosis. These results suggest that compromised autophagy may contribute to the development of OA.
MATERIALS AND METHODS
Human cartilage procurement and processing
Normal human articular cartilage from 14 adult donors (mean ± SD 22.4 ± 6.2 years; Mankin score = 0, OA grade I) having no history of joint disease (inflammatory arthritis, OA, microcrystalline arthritis, or osteonecrosis) was harvested from femoral condyles and tibial plateaus at autopsy. Human cartilage was also obtained at autopsy from 8 older donors having no history of joint diseases or overt OA (mean ± SD 69.5 ± 9.02, Mankin score = 3, OA grade II) but mild aging-related changes. Osteoarthritic human articular cartilage was obtained from 16 patients (mean ± SD 74.7 ± 9.4 years; Mankin score = 7–8, OA grade III–IV) undergoing knee replacement surgery. Human tissues were obtained under approval by the Scripps Human Subjects Committee. All tissue samples were graded macroscopically according to a modified Outerbridge scale (23) and Safranin O stained sections of normal and osteoarthritic cartilage were graded according to Mankin (24): Mankin score 0, normal cartilage; Mankin score 1–4, mild osteoarthritic cartilage; and Mankin score ≥ 5, osteoarthritis cartilage (25). Once cartilage surfaces were rinsed with saline, scalpels were used to cut parallel sections 5 mm apart, vertically from the cartilage surface onto the subchondral bone. These cartilage strips were then resected from the bone. Human chondrocytes were isolated and cultured as described previously (26). The cartilage tissue was incubated with trypsin (0.5 mg/ml; Sigma-Aldrich, St. Louis, MO) at 37°C for 10 minutes. After the trypsin solution was removed, the tissue slices were treated for 12 to 16 hours with type IV clostridial collagenase (2 mg/ml, Sigma-Aldrich) in Dulbecco's modified Eagle's medium (DMEM, Mediatech, Manassas, VA) with 5% fetal calf serum.
Primary culture of chondrocytes
The isolated chondrocytes were recovered and plated at high density in Dulbecco’s modified Eagle’s (high glucose) medium supplemented with 10% calf serum (CS), L-glutamine, and antibiotics and allowed to attach to the culture flasks. The cells were incubated at 37°C in a humidified gas mixture containing 5% CO2 balanced with air. The chondrocytes were used in the experiments at confluency (2–3 weeks in primary culture).
Experimental osteoarthritis models in mice
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) at The Scripps Research Institute. In the spontaneous aging-related OA model, C57BI/6J mice were kept under normal conditions and knee joints were compared at 2, 9 and 12 months of age. The surgical OA model was induced in 2 months old C57Bl/6J mice by transection of the medial meniscotibial ligament and the medial collateral ligament (MMTL+MCL) and animals were euthanized 8 weeks later. Knee joints from both OA models were resected from both hind legs, fixed in 10% zinc-buffered formalin (Z-Fix; Anatech, Battle Creek, MI) for 2 days, decalcified in TBD-2 (Shandon, Pittsburgh, PA) for 24 hours, followed by paraffin embedding. Serial sections (4 µm each) were cut, stained with Safranin O-fast green, and examined for histopathological changes. ULK1, Beclin1, LC3 and PARP p85 expression was analyzed by immunohistochemistry in the two different OA animal models.
Quantification and localization of positive cells in human cartilage
ULK1, Beclin1 and LC3 localization in each cartilage zone was assessed systematically by counting positive cells in 50 × 50 µm grids starting from the cartilage surface to the deep zone. This was repeated a minimum of three times for each section. The identification of each zone was based on previously reported characteristics that comprise cell shape, morphology, orientation, and pericellular matrix (PM) deposition (27). The depth of each zone was recorded for each section for comparative analysis on the frequency of positive cells in each zone. The frequency of positive cells was expressed as a percentage relative to the total number of cells counted in each zone.
Quantification of cartilage positive cells in mouse models
Cartilage cellularity in C57BL/6J WT mice was quantified by counting the chondrocytes in a microscopic field (28). In cartilage from aging-related OA model (2, 9 and 12-month-old mice) and surgically-induced OA (Sham surgery and 8 weeks after surgery), 3 pictures were taken under 40X magnification, representing the center of the femoral condyle that is not covered by the menisci as well as the medial and lateral femoral condyles. Then, the total number of ULK1, Beclin1 and LC3-positive cells was counted in each section and was expressed as the percentage relative to the 2 months old mice (control) and Sham (control) in aging-related OA and surgically-induced OA, respectively.
Western blotting
Cells were washed in ice-cold PBS, pH 7.5, and lysed in 0.2 M TrisHCl, pH 6.8, containing 2% sodium dodecyl sulfate (SDS), 20% glycerol, 1 mg/ml protease inhibitor cocktail (SigmaAldrich) and 1 mM phenyl methyl sulfonyl fluoride (PMSF, SigmaAldrich). Whole cell lysates were boiled for 5 min. Cartilage was cut into 1 mm thin slices and 200 to 1000 mg of frozen cartilage was pulverized in a liquid nitrogen-cooled freezer-mill for two cycles of 1.5 min at the rate of maximum impact frequency. Dry weight of normal and OA cartilage was measured and the same amount of protein was resuspended in SDS gel loading buffer (50mM Tris PH 6.8, 10% glycerol, 4% sodium dodecyl sulfate, 10% 2-mercaptoethanol, and 0.001% bromophenol blue) and mixed for 2 hours at room temperature. Centrifugation at 14000-rpm was performed for 30 minutes, and then supernatants were harvested and heated 80 for 10 minutes. The protein concentrations were determined using a bicinchoninic acid (BCA) reagent assay (Pierce Chemical, Rockford, IL). The concentrated samples were then adjusted to equal volumes before resolution on 12% Tris-Glycine gels (Invitrogen, Carlsbad, CA). Protein was transferred to nitrocellulose membranes (Invitrogen), blocked with 5% dry milk in Tris-buffered saline–Tween (TBST), and blotted with rabbit polyclonal antibody specific for ULK1 and Beclin1 (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit polyclonal antibody for LC3 (Abgent, San Diego, CA) and Glyceraldehyde-3-phosphate dehydrogenase antibody (GAPDH) (Ambion, Austin, TX) for one hour. The membranes were then incubated with HRP-conjugated anti-mouse IgG (Cell Signalling Technology, Boston, MA) for one hour. Afterwards, the membranes were washed three times with TBST and developed using the enhanced chemiluminescent substrate from Pierce (Rockford, IL).
Immunohistochemistry
Paraffin-embedded samples were first deparaffinized in the xylene substitute Pro-Par Clearant (Anatech) and rehydrated in graded ethanol and water. For antigen unmasking, sections in 10 mM sodium citrate buffer (pH 6.0) were heated in a microwave oven and kept at 80–85°C for 1.5 minutes. Slides were cooled for 20 minutes at room temperature after antigen unmasking. After washing with phosphate buffered saline (PBS), sections were blocked with 5% serum for 30 minutes at room temperature. The following steps were performed: ULK1 antibody (1:100 dilution), Beclin1 antibody (1:100 dilution), LC3 (1:100 dilution) and negative control IgG (1 µg/ml) were applied and incubated overnight at 4°C. After washing with PBS, sections were treated with 3% hydrogen peroxide (H2O2) for 10 minutes, washed with PBS, incubated with biotinylated goat anti-rabbit secondary antibody for 30 minutes at room temperature, and then incubated using the Vectastain ABC-AP kit (Vector Laboratories, Burlingame, CA) for 30 minutes. Slides were washed, and sections were incubated with 3,3-diaminobenzidine tetrahydrochloride (DAB) substrate for 3–10 minutes.
Detection of poly (ADP-ribose) polymerase (PARP) cleavage
Polyclonal antibody specific for the p85 fragment of PARP (Promega, Madison, WI) was used. Sections were microwaved in PBS–citrate buffer, pH 6.0, then digested with hyaluronidase, washed, and blocked for 30 min in PBS containing 0.1% Tween 20 and 10% normal serum. Primary antibody was applied at a 1:200 dilution and incubated overnight at 4°C. The following day, the sections were washed, blocked with 3% H2O2 for 5 min, washed again, and incubated with diluted secondary antibody (HRP-conjugated anti-rabbit IgG) for 1 hour. The slides were incubated for 30 min with peroxidase-based Elite ABC system (Vector Laboratories), and then incubated for 4–10 min in DAB substrate, followed by mounting as described above.
Statistical analysis
Statistically significant differences between two groups were determined with t- tests. The results are reported as mean ± S.D. P values less than 0.05 were considered significant.
RESULTS
ULK1, Beclin1 and LC3 expression in human articular cartilage
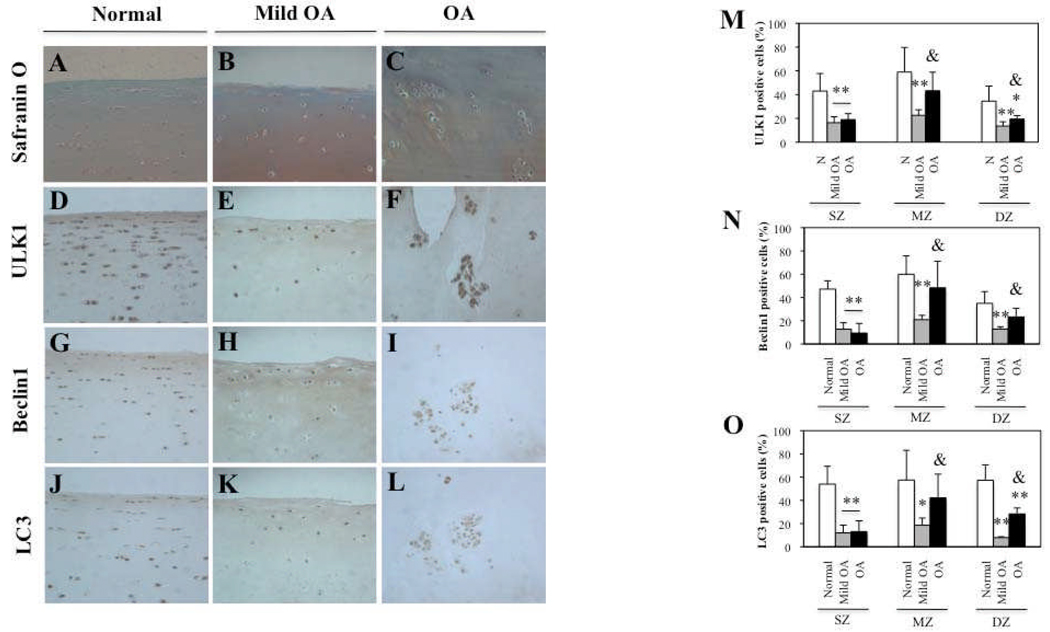
To study the role of autophagy in the development of osteoarthritis (OA), we evaluated the expression of ULK1, an inducer of autophagy, Beclin1, a regulator of autophagy and LC3, which executes autophagy, in human Normal (Mankin score= 0, OA grade I), Mild osteoarthritic (OA) (Mankin score= 3, OA grade II), and OA (Mankin score= 7–8, OA grade III–IV), articular cartilage. Cartilage samples were evaluated by staining with Safranin O (Figure 1A–C) and ULK1, Beclin1 and LC3 expression was analyzed by immunohistochemistry (IHC) (Figure 1D–L). The quantitative analysis of distribution of positive cells (Figure 1M,N,O) showed that in Normal cartilage, ULK1 (mean ± SD 23.2 ± 9.7 years, n=5), Beclin1 and LC3 (mean ± SD 24.5 ± 9.0 years, n=6) were highly expressed in superficial zone (SZ), middle zone (MZ) and deep zone (DZ) (Figure 1D,G,J). In Mild OA, ULK1, Beclin1 and LC3 (mean ± SD 74.5 ± 7.0 years, n=4) expression were significantly decreased in the SZ (P < 0.01), MZ (P < 0.01 and P < 0.05) and DZ (P < 0.01) compared to Normal cartilage. In OA cartilage, ULK1 (mean ± SD 71 ± 10.7 years, n=5), Beclin1 and LC3 (mean ± SD 74.7 ± 5.5 years, n=6) expression were reduced in all zones compared to Normal cartilage. This reduction was significant in the SZ for ULK1, Beclin1 and LC3 (P < 0.01) and in DZ for ULK1 and LC3 (P < 0.05 and P < 0.01, respectively). In contrast, ULK1, Beclin1 and LC3 expression were significantly increased in MZ (P < 0.05) and DZ (P < 0.05) compared to mild OA. This increase was due to the cell clusters localized in MZ and DZ in OA cartilage. These cell clusters showed strong expression of ULK1, Beclin1 and LC3 (Figure 1F,I,L).
Figure 1.
ULK1, Beclin1 and LC3 expression is reduced in human osteoarthritic (OA) cartilage. A–C, Representative sections of 6 normal (N) cartilage, 4 mild OA and 6 OA cartilage samples as shown by Safranin O. D–F, G–I, J–L, Immunohistochemical analysis with anti-ULK1, Beclin1 and LC3, respectively. M–O, Quantification of ULK1, Beclin1 and LC3-positive cells in N, mild OA and OA cartilage. In mild OA, the percentage of ULK1, Beclin1 and LC3-positive cells was significantly reduced in superficial (SZ), middle (MZ) and deep zone (DZ) compared to N cartilage. Values are the mean ± SD. ** = P < 0.01 versus N cartilage; * = P < 0.05 versus N cartilage. In OA cartilage, ULK1, Beclin1 and LC3 expression were significantly reduced in SZ compared to N cartilage. In DZ only ULK1 and LC3 expression was significantly reduced. Values are the mean ± SD. ** = P < 0.01 versus N cartilage; * = P < 0.05 versus N cartilage in DZ. Furthermore, ULK1, Beclin1 and LC3 expression was significantly increased in OA cartilage compared to mild OA in MZ and DZ. Values are the mean ± SD. & = P < 0.05 versus Mild OA. Magnification: A–L x40.
These results indicate that ULK1, Beclin1 and LC3 are expressed in Normal cartilage with an overall reduction in mild OA and OA cartilage and high expression in the OA cluster chondrocytes.
ULK1, Beclin1 and LC3 protein expression in human cultured articular chondrocytes
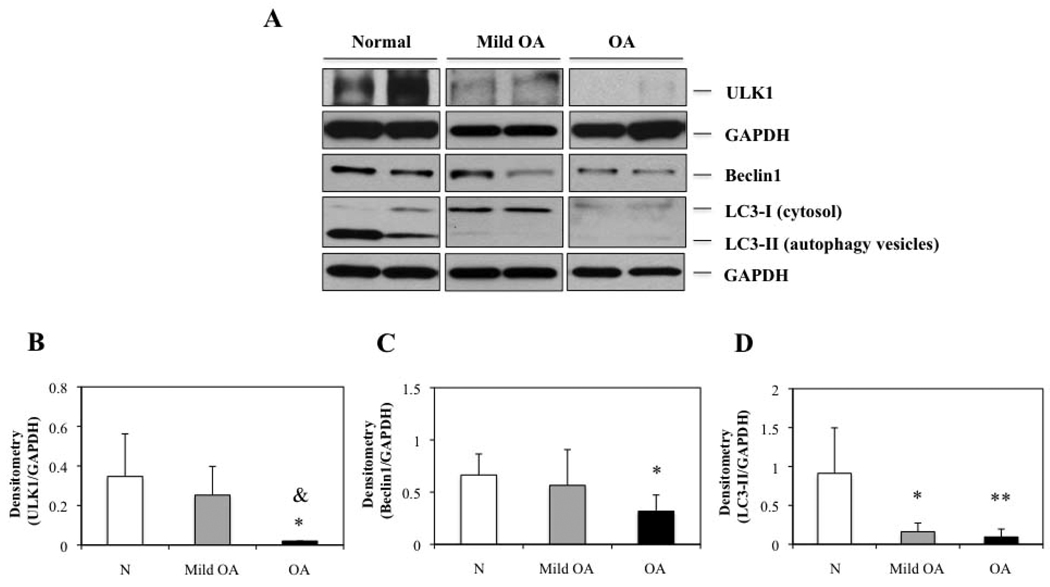
To confirm the results obtained in cartilage, we evaluated ULK1, Beclin1 and LC3 protein expression by Western blotting in chondrocytes from Normal donors (mean ± SD 20.6 ± 3.1 years for ULK1 and mean ± SD 21.4 ± 3.4 years for Beclin1 and LC3), Mild OA donors (mean ± SD 64.5 ± 8.5 years) and OA donors (mean ± SD 71.5 ± 9.1 years for ULK1 and mean ± SD 76.6 ± 10.9 years for Beclin1 and LC3) (Figure 2A). ULK1, Beclin1 and LC3-II, an indicator of autophagosome formation were expressed in normal chondrocytes and were decreased in mild OA and OA chondrocytes. Densitometric analysis showed a significant decrease of 17-fold, 2.1-fold and 9-fold for ULK1, Beclin1 and LC3-II, respectively compared to Normal chondrocytes (P < 0.05 and P < 0.01) (Figure 2C–D) (Figure 2B–D). In addition, ULK1 protein expression was significantly reduced in OA chondrocytes compared to Mild OA (P < 0.05) (Figure 2A). These results suggest that autophagy inducer ULK1, autophagy regulator Beclin1 and autophagy marker LC3, show similar expression patterns in Normal cartilage and chondrocytes and similar abnormal expression in OA. Importantly, LC3-II which indicates active autophagy in Normal tissue but a reduction in OA.
Figure 2.
ULK1, Beclin1 and LC3 expression is decreased in human osteoarthritic (OA) chondrocytes. A, Total protein from N, mild OA and OA chondrocytes was analyzed by Western blotting using anti-ULK1, Beclin1, LC3 and GAPDH as described in methods. The images shown are representative of 4 N, mild OA and OA donors for ULK1 and 6 N, 4 mild OA and 6 OA donors for Beclin1 and LC3. Representative blots are shown along with the numeric data obtained by densitometry. B–D, Densitometry analysis showed a significant decreased of protein expression by 17-fold, 2.1-fold and 9-fold for ULK1, Beclin1 and LC3-II, respectively, in OA chondrocytes compared to N chondrocytes. Values are the mean ± SD. * = P < 0.05 versus N cartilage and ** = P < 0.01 versus N cartilage. On the other hand, ULK1 protein expression was significantly reduced in OA chondrocytes compared to mild OA. Values are the mean ± SD. & = P < 0.05 versus mild OA.
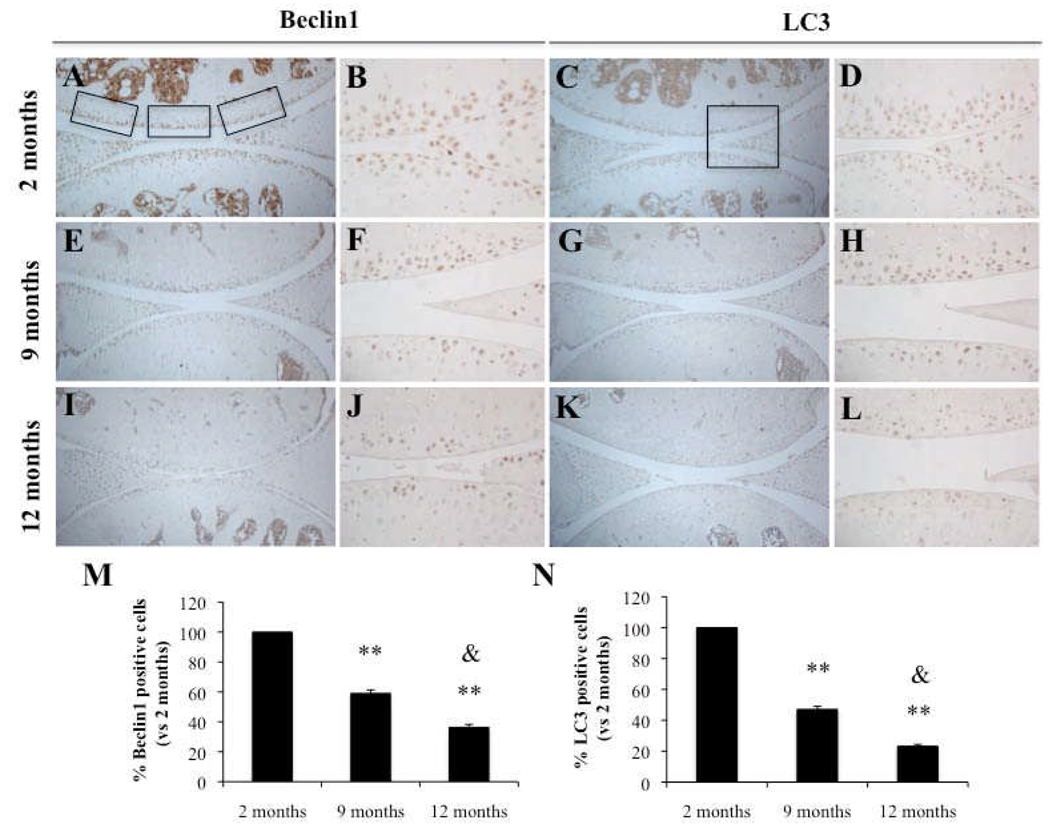
Aging-related cartilage degradation and reduction in Beclin1 and LC3 expression in mouse articular cartilage
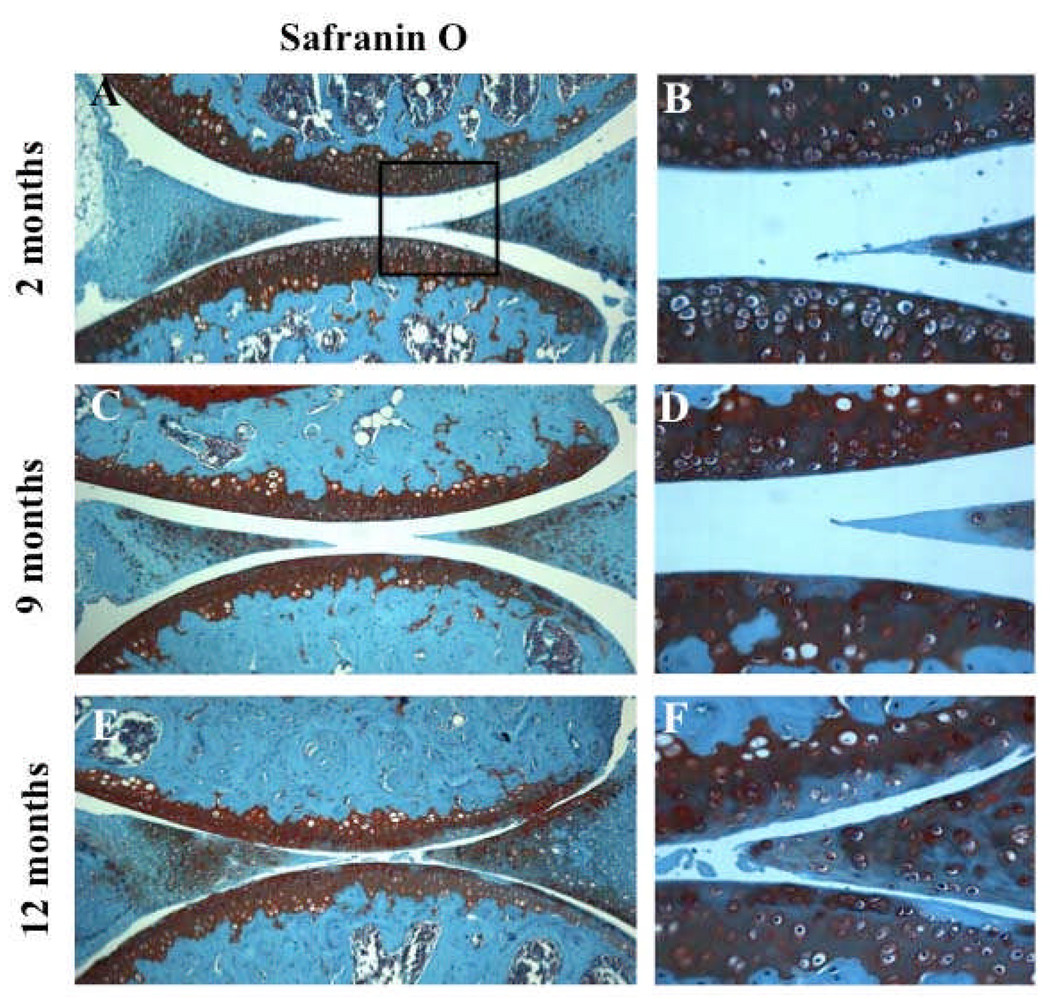
Articular cartilage from 2-month old C57Bl/6J WT mice showed homogeneous Safranin O staining in all zones, intact surface, and high cell density (Figure 3A–B). At 9 months, C57Bl/6J WT developed mild OA-like changes as previously reported (29) with modest reduction in Safranin O staining (Figure 3C–D). At 12 months, C57Bl/6J WT developed more obvious OA changes with moderate reduction of Safranin O staining and reduction in cartilage thickness and cellularity (Figure 3E–F). Immunohistochemical analysis of knee joints from C57BL/6J mice aged 9 and 12 months showed an aging-related significant reduction in Beclin1 and LC3-positive cells (P < 0.01) compared to 2 month old animals and significant reduction in Beclin1 and LC3 at 12 months compared to 9 months old animals (P < 0.05) (Figure 4M–N). At 2 months, Beclin1 and LC3 were highly expressed in the SZ and upper MZ (Figure 4A–D); however, this expression was reduced at 9 and 12 months when OA-like changes had developed (Figure 4E–H; Figure 4I–L).
Figure 3.
Aging-related loss of glycosaminoglycans (GAGs) in mouse joints. A–F, Knee joints from 2 months (n=4), 9 months (n=4) and 12 months (n=4) old C57Bl/6J mice were analyzed by Safranin O staining. Magnification: A,C,E x10; B,D,F x40.
Figure 4.
Aging-related reduction in Beclin1 and LC3 expression in mouse joints. Knee joints from 2, 9 and 12 month old C57Bl/6J mice were analyzed by immunohistochemistry (IHC) for Beclin1 (A–B, E–F, I–J) and LC3 (C–D, G–H, K–L). M–N, Quantification of Beclin1 and LC3-positive cells at 2, 9 and 12 months. Beclin1 and LC3-positive cells were significantly decreased at 9 and 12 months compared to 2 months. Values are the mean ± SD. ** = P < 0.01 versus 2 months old. Furthermore, Beclin1 and LC3-positive cells were significantly reduced at 12 months compared to 9 months. Values are the mean ± SD. & = P < 0.05 versus 9 months old. Knee joints from C57BL/6JWT mice age 2 months (n=4), 9 months (n=4) and 12 months (n=4) were examined. Magnification: A,C,E,G,I,K x10; B,D,F,H,J,L x40.
Reduction in ULK1 and Beclin1 expression in mouse joints with surgically-induced OA
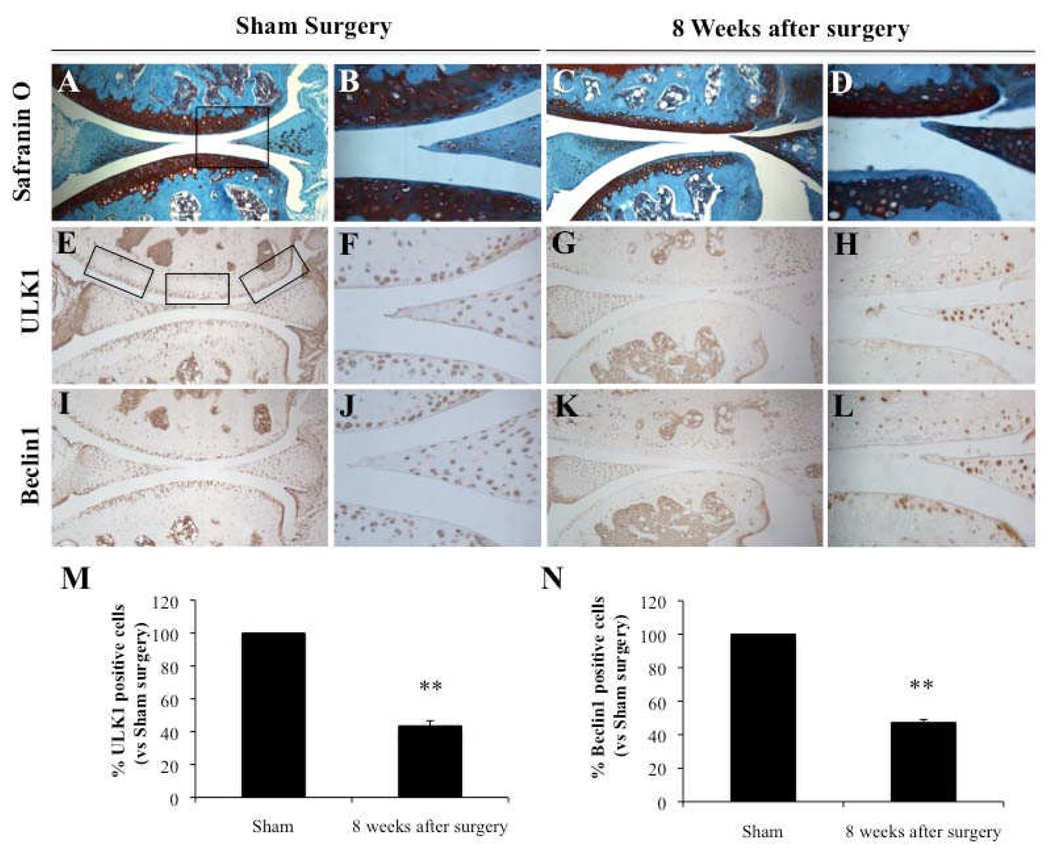
OA was induced in knee joints from 2 months old C57Bl/6J mice by transection of the medial meniscotibial ligament and the medial collateral ligament (MMTL+MCL). Knee joints at 8 weeks after surgery showed OA pathology (Figure 5C–D). Immunohistochemical analysis of knee joints at that time point showed a reduction of ULK1 and Beclin1-positive cells (Figure 5G–H; Figure 5K–L) compared to control knees with sham surgery (Figure 5E–F; Figure 5I,J). The quantitative analysis of positive cells showed a significant reduction of ULK1 and Beclin1 at 8 weeks after surgery (P < 0.01) (Figure 5 M–N).
Figure 5.
Reduction in ULK1 and Beclin1 expression in knee joints from mice with surgical OA. Knee joints from 2 months old C57Bl/6J mice with Sham surgery (n=4) and with surgical OA induced by transection of the medial meniscotibial ligament and the medial collateral ligament (MMTL+MCL) at 8 weeks (n=4) were analyzed by Safranin O staining (A–D) or for ULK1 (E–H) and Beclin1 (I–L). M–N, Quantification of ULK1 and Beclin1-positive cells. The expression was significantly decreased at 8 weeks after surgery. Values are the mean ± SD. ** = P < 0.01 versus Sham surgery. Magnification: A,C,E,G,I,K x10; B,D,F,H,J,L x40.
Increased cell death in aging-related and surgical OA mouse joints
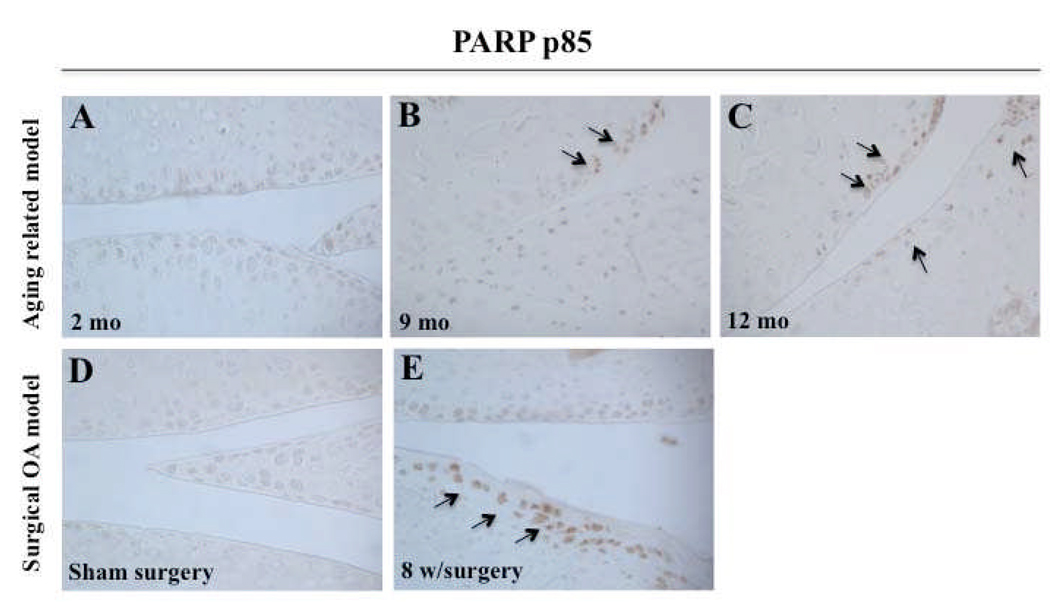
To further investigate the relationship between autophagy and apoptotic cell death, immunohistochemistry was performed for the apoptosis marker poly (ADP-ribose) polymerase (PARP) p85 on the same samples that were analyzed in the preceding experiments for the autophagy markers. In knee joints from 9 and 12 month old C57Bl/6J mice, significantly more chondrocytes were positive for PARP-p85 as compared to 2 month old mice (Figure 6A–C). In knee joints with surgically induced OA, an increased number of PARP p85-positive cells was observed at 8 weeks after surgery as compared to control (Figure 6D–E). Together, these findings demonstrate that human ageing and OA cartilage and chondrocytes, aging-related and surgical OA mice models are associated with a reduction in autophagy (ULK1, Beclin1 and LC3 expression) along with an increase of apoptosis (PARP p85).
Figure 6.
Increased cell death in aging-related and surgical OA in mouse joints. A–C, Knee joints from 2, 9, 12 month old C57Bl/6J mice were used to evaluate PARP p85 expression by immunohistochemistry (IHC). Knee joints from C57BL/6JWT mice aged 2 months (n=3), 9 months (n=3) and 12 months (n=3) were examined. D–E, Knee joints from 2 months old C57Bl/6J mice with sham surgery (n=3) and surgical OA at 8 weeks (n=3) were evaluated for PARP p85 expression by IHC. Magnification: x40.
DISCUSSION
The relationship between aging and osteoarthritis is clinically and epidemiologically evident and recent findings provide insight into mechanisms that lead to aging-related changes in cells and extracellular matrix (30). Articular cartilage appears to be highly susceptible to the accumulation of aging-related changes, in part due to the relatively low turnover of extracellular matrix and cells. It has been estimated that the half-life of type II collagen is approximately 117 years (31) and various measurements in mature cartilage revealed only a very small fraction of proliferating cells (32). Thus, cartilage matrix and cells are prone to accumulate changes related to trauma, mechanical or oxidant stress over time (33). At the cellular level, a large number of aging-related changes have been documented. These include an overall reduction in cartilage cellularity, reduced antioxidant defenses, altered responses to growth factors and cytokines and changes in gene and protein expression patterns (21). Physiological mechanisms that allow for repair of cellular damage are thus critical for the maintenance of chondrocyte survival and function. Best characterized in this regard are mechanisms for nuclear and mitochondrial DNA repair. In contrast, mechanisms capable of replacing damaged proteins and organelles in chondrocytes are essentially unknown.
Autophagy has gained interest in the past decade due to its role in the pathogenesis of various diseases and in regulation of the aging process. Autophagy has been characterized as a highly regulated cellular mechanism with both beneficial and pathogenic effects (34). Normal cellular development and growth require a well-regulated balance between protein synthesis and degradation. Eukaryotic cells have two major mechanisms for degradation, the proteasome and autophagy pathways. Autophagy is involved in the bulk degradation of long-lived cytosolic proteins and organelles, whereas the ubiquitin proteasome system degrades specific short-lived proteins (35). Recent studies provide compelling evidence that at least in model organisms autophagy protects against diverse pathologies, such as, neurodegeneration, heart diseases, infections, cancer and aging (8). Genetic studies in mice support the importance of autophagy in physiological and pathological events. In fact, loss of autophagy genes leads to neurodegeneration, cardiomyophaties and abnormalities in skeletal development and is associated with the accumulation of cytoplasmic protein aggregates (18,36,37,38,39). The mechanisms by which autophagy is lost with aging are mainly related to the failure of the lysosomal hydrolases, resulting in an increase of toxic protein products and slow clearance of autophagosomes in the aging tissues (40). In addition, other studies described alterations in the response of macroautophagy to hormonal changes with aging. In particular, the effects of oxidative stress on the insulin receptor-signaling pathway seem to play a critical role in decreased autophagy in aged organisms (41). The signaling network involving longevity factors SIRT1, mTOR, FoxO3, NF-κB and p53 regulate autophagy and might have a role in the aging process. mTOR and NF-κB are repressors of the autophagy pathway under input signals of stress and inflammation, while SIRT1, a stress resistance and longevity factor, and FoxO3, a major regulator of cellular metabolism, proliferation and stress resistance, enhance autophagy (42).
The present study demonstrates that ULK1, Beclin1 and LC3 are expressed in normal and murine articular cartilage, suggesting activation of autophagy and the presence of LC3-II directly indicates autophagosome formation (14). However, in OA cartilage and chondrocytes, the expression of these autophagy markers was significantly decreased. Importantly, the reduction in LC3-II implies defective or reduced autophagy in OA. These observations are consistent with the notion that the basal autophagic activity decreases with the age, thus contributing to the accumulation of damaged macromolecules and susceptibility to aging-related diseases (43). Abnormal protein aggregation and formation of characteristic pathological structures are central features of such diseases. Interestingly, cellular responses to protein misfolding and aggregation are intimately related to mechanisms of pathogenesis and potential targets of new therapies (44).
The present results also show that autophagy was decreased in the surgical OA model in mice. Since this was observed in relatively young mice (2–4 month old) it is apparently not a consequence of aging-related mechanisms. We found similar results in porcine cartilage explants following exposure to mechanical injury (unpublished). It will be of interest to determine mechanisms that are responsible for the suppression or loss of autophagy in the surgical OA model and after mechanical injury. However, both the surgical OA model and mechanically injured cartilage feature increased cell death suggesting that loss of autophagy may contribute to cell death.
Little is known about the role of autophagy in articular cartilage. Morphological changes similar to autophagic cell death have been described (45). More information is available on autophagy studies in the epiphyseal growth plate. Autophagy regulates maturation and promotes survival of terminally differentiated chondrocytes under stress and hypoxia conditions (46,47,48). In fact, silencing the transcription factor hypoxia-inducible factor 1 (HIF-1) in chondrocytes, decreases the expression of Beclin1, suggesting a role of this factor as a positive autophagy regulator and that it could play a protective role against cell death (48). By contrast, suppression of HIF-2 in OA and aging cartilage was associated with increased HIF-1 expression and autophagy induction (49).
The protective role of autophagy in endochondral ossification was further supported by observations that its inactivation leads to severe skeletal abnormalities, due in part to cell death (39). In addition, autophagy is essential for early embryonic development, supported by observations that Beclin1 disruption causes early embryonic lethality, autophagy deficiency and apoptosis (50). In order to investigate the relationship between autophagy and apoptotic cell death as well as the mechanism of chondrocyte loss and cartilage degradation in aging and OA, immunohistochemistry was performed in the present study for the apoptosis marker poly (ADP-ribose) polymerase (PARP) p85. We found that in the mouse aging-related and surgically-induced OA models autophagy is decreased, while apoptotic cell death is increased. This correlation requires further analysis to determine whether a direct causal relationship exists between the two processes in cartilage.
In summary, this study is the first to demonstrate that autophagy may be a protective or homeostatic mechanism in normal cartilage. By contrast, human OA and spontaneous and surgically-induced OA in mice are associated with a reduction and loss of ULK1, Beclin1 and LC3 expression and a related increase in apoptosis. These results suggest that compromised autophagy may represent a novel mechanism in the development of OA.
Acknowledgements
We are thankful to Jean Valbracht and Lilo Creighton for technical assistance.
This study was supported by National Institutes of Health Grants AG007996, AG033409 and the Sam and Rose Stein Endowment Fund. B. Caramés was supported by Postdoctoral Fellowship “Anxeles Alvariño”, Secretaria Xeral I+D+i, Xunta de Galicia, Spain. N. Taniguchi was supported by an award from the Arthritis National Research Foundation (ANRF). S. Otsuki was supported by a postdoctoral fellowship from the Arthritis Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
Dr. Lotz had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study design: Lotz, Blanco, Caramés
Acquisition of data: Caramés, Taniguchi, Otsuki
Analysis and interpretation of data: Caramés, Lotz
Manuscript preparation: Lotz, Blanco, Caramés
Statistical analysis: Caramés
REFERENCES
- 1.Ruiz-Romero C, Calamia V, Mateos J, Carreira V, Martínez-Gomariz M, Fernández M, et al. Mitochondrial dysregulation of osteoarthritic human articular chondrocytes analyzed by proteomics: A decrease in mitochondrial superoxide dismutase points to a redox imbalance. Mol Cell Proteomics. 2009;8:172–189. doi: 10.1074/mcp.M800292-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taniguchi N, Caramés B, Ronfani L, Ulmer U, Komiya S, Bianchi ME, et al. Aging-related loss of the chromatin protein HMGB2 in articular cartilage is linked to reduced cellularity and osteoarthritis. Proc Natl Acad Sci U S A. 2009;27(106):1181–1186. doi: 10.1073/pnas.0806062106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pennock AT, Robertson CM, Emmerson BC, Harwood FL, Amiel D. Role of apoptotic and matrix-degrading genes in articular cartilage and meniscus of mature and aged rabbits during development of osteoarthritis. Arthritis Rhem. 2007;56:1529–1536. doi: 10.1002/art.22523. [DOI] [PubMed] [Google Scholar]
- 4.Aigner T, Haag J, Martin J, Buckwalter J. Osteoarthritis: aging of matrix and cells--going for a remedy. Curr Drug Targets. 2007;8:325–331. doi: 10.2174/138945007779940070. [DOI] [PubMed] [Google Scholar]
- 5.Vellai T. Autophagy genes and ageing. Autophagy genes and ageing. Cell Death Differ. 2009;16:94–102. doi: 10.1038/cdd.2008.126. [DOI] [PubMed] [Google Scholar]
- 6.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol. 2004;36:2445–2462. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Thorburn A. Apoptosis and autophagy: regulatory connections between two supposedly different processes. Apoptosis. 2008;13:1–9. doi: 10.1007/s10495-007-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator ofautophagy. J Biol Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 12.Furuya N, Yu J, Byfield M, Pattingre S, Levine B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005;1:46–52. doi: 10.4161/auto.1.1.1542. [DOI] [PubMed] [Google Scholar]
- 13.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12:1509–1518. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 17.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, et al. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci U S A. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 20.Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldring MB, Goldring SR. Osteoarthritis. J Cell Physiol. 2007;213:626–634. doi: 10.1002/jcp.21258. [DOI] [PubMed] [Google Scholar]
- 22.Cuervo AM. Autophagy and aging: keeping that old broom working. Trends Genet. 2008;24:604–612. doi: 10.1016/j.tig.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamada K, et al. Subchondral bone of the human knee joint in aging and osteoarthritis. Osteoarthritis Cartilage. 2002;10:360–369. doi: 10.1053/joca.2002.0525. [DOI] [PubMed] [Google Scholar]
- 24.Mankin HJ, Dorfman H, Lippiello L, Zarins A. Biochemical and metabolic abnormalities in articular cartilage from osteo-arthritic human hips. II. Correlation of morphology with biochemical and metabolic data. J Bone Joint Surg Am. 1971;53:523. [PubMed] [Google Scholar]
- 25.Pfander D, Cramer T, Weseloh G, Pullig O, Schuppan D, Bauer M, et al. Hepatocyte growth factor in human osteoarthritic cartilage. Osteoarthritis Cartilage. 1999;7:548–559. doi: 10.1053/joca.1999.0259. [DOI] [PubMed] [Google Scholar]
- 26.Blanco FJ, Ochs RL, Schwarz H, Lotz M. Chondrocyte apoptosis induced by nitric oxide. Am J Pathol. 1995;146:75–85. [PMC free article] [PubMed] [Google Scholar]
- 27.Guilak F, Alexopoulos LG, Upton ML, Youn I, Choi JB, Cao L, et al. The pericellular matrix as a transducer of biomechanical and biochemical signals in articular cartilage. Ann N Y Acad Sci. 2006;1068:498–512. doi: 10.1196/annals.1346.011. [DOI] [PubMed] [Google Scholar]
- 28.Zemmyo M, Meharra EJ, Kühn K, Creighton-Achermann L, Lotz M. Accelerated, aging-dependent development of osteoarthritis in alpha1 integrin-deficient mice. Arthritis Rheum. 2003;48:2873–2880. doi: 10.1002/art.11246. [DOI] [PubMed] [Google Scholar]
- 29.Stanescu R, Knyszynski A, Muriel MP, Stanescu V. Early lesions of the articular surface in a strain of mice with very high incidence of spontaneous osteoarthritic-like lesions. J Rheumatol. 1993;20:102–110. [PubMed] [Google Scholar]
- 30.Aigner T, Söder S, Gebhard PM, McAlinden A, Haag J. Mechanisms of disease: role of chondrocytes in the pathogenesis of osteoarthritis-structure, chaos and senescence. Nat Clin Pract Rheumatol. 2007;3:391–399. doi: 10.1038/ncprheum0534. [DOI] [PubMed] [Google Scholar]
- 31.Rötzer A, Mohr W. 3H-thymidine incorporation into chondrocytes of arthritic cartilage. Z Rheumatol. 1992;51:172–176. [PubMed] [Google Scholar]
- 32.Verzijl N, DeGroot J, Thorpe SR, Bank RA, Shaw JN, Lyons TJ, et al. Effect of collagen turnover on the accumulation of advanced glycation end products. J Biol Chem. 2000;275:39027–39031. doi: 10.1074/jbc.M006700200. [DOI] [PubMed] [Google Scholar]
- 33.Martin JA, Brown T, Heiner A, Buckwalter JA. Post-traumatic osteoarthritis: the role of accelerated chondrocyte senescence. Biorheology. 2004;41:479–491. [PubMed] [Google Scholar]
- 34.Dwivedi M, Ahnn J. Autophagy-Is it a preferred route for lifespan extension? BMB Rep. 2009;42:62–71. [PubMed] [Google Scholar]
- 35.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12:1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 38.Shibata M, Lu T, Furuya T, Degterev A, Mizushima N, Yoshimori T, et al. Regulation of intracellular accumulation of mutant Huntingtin by Beclin1. J Biol Chem. 2006;281:14474–14485. doi: 10.1074/jbc.M600364200. [DOI] [PubMed] [Google Scholar]
- 39.Settembre C, Arteaga-Solis E, McKee MD, de Pablo R, Al Awqati Q, Ballabio A, et al. Proteoglycan desulfation determines the efficiency of chondrocyte autophagy and the extent of FGF signaling during endochondral ossification. Genes Dev. 2008;22:2645–2650. doi: 10.1101/gad.1711308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brunk UT, Terman A. Lipofuscin: mechanisms of age-related accumulation and influence on cell function. Free Radic Biol Med. 2002;33:611–619. doi: 10.1016/s0891-5849(02)00959-0. [DOI] [PubMed] [Google Scholar]
- 41.Cuervo AM, Bergamini E, Brunk UT, Dröge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining "clean" cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- 42.Salminen A, Kaarniranta K. Regulation of the aging process by autophagy. Trends Mol Med. 2009;15:217–224. doi: 10.1016/j.molmed.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 43.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 44.Kim DH, Davis RC, Furukawa R, Fechheimer M. Autophagy contributes to degradation of Hirano bodies. Autophagy. 2009;5:44–51. doi: 10.4161/auto.5.1.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roach HI, Aigner T, Kouri JB. Chondroptosis: a variant of apoptotic cell death in chondrocytes? Apoptosis. 2004;9:265–277. doi: 10.1023/b:appt.0000025803.17498.26. [DOI] [PubMed] [Google Scholar]
- 46.Srinivas V, Bohensky J, Shapiro IM. Autophagy: a new phase in the maturation of growth plate chondrocytes is regulated by HIF, mTOR and AMP kinase. Cells Tissues Organs. 2009;189:88–92. doi: 10.1159/000151428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srinivas V, Shapiro IM. Chondrocytes embedded in the epiphyseal growth plates of long bones undergo autophagy prior to the induction of osteogenesis. Autophagy. 2006;2:215–216. doi: 10.4161/auto.2649. [DOI] [PubMed] [Google Scholar]
- 48.Bohensky J, Shapiro IM, Leshinsky S, Terkhorn SP, Adams CS, Srinivas V. HIF-1 regulation of chondrocyte apoptosis: induction of the autophagic pathway. Autophagy. 2007;3:207–214. doi: 10.4161/auto.3708. [DOI] [PubMed] [Google Scholar]
- 49.Bohensky J, et al. Regulation of autophagy in human and murine cartilage: hypoxia-inducible factor 2 suppresses chondrocyte autophagy. Arthritis Rheum. 2009;60:1406–1415. doi: 10.1002/art.24444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]