Abstract
It is of fundamental importance to understand the determinants of the rate of protein evolution. Eukaryotic extracellular proteins are known to evolve faster than intracellular proteins. Although this rate difference appears to be due to the lower essentiality of extracellular proteins than intracellular proteins in yeast, we here show that, in mammals, the impact of extracellularity is independent from the impact of gene essentiality. Our partial correlation analysis indicated that the impact of extracellularity on mammalian protein evolutionary rate is also independent from those of tissue-specificity, expression level, gene compactness, and the number of protein–protein interactions and, surprisingly, is the strongest among all the factors we examined. Similar results were also found from principal component regression analysis. Our findings suggest that different rules govern the pace of protein sequence evolution in mammals and yeasts.
Keywords: evolutionary rate, subcellular localization, gene essentiality, gene expression level, mammal, yeast
It has been of great interest among molecular evolutionists to identify factors that explain the large variation in the evolutionary rate of proteins encoded in a genome (Fraser et al. 2002; Subramanian and Kumar 2004; Drummond et al. 2005; Zhang and He 2005; Liao et al. 2006; Makino and Gojobori 2006). Extracellular proteins, also known as secreted proteins, have been shown to exhibit elevated rates of nonsynonymous substitutions in both yeasts and mammals (Winter et al. 2004; Julenius and Pedersen 2006; Dean et al. 2008), even after the control of gene expression level and number of protein interactions (Julenius and Pedersen 2006). In yeast, however, the evolutionary rate is no longer significantly different between extra- and intracellular proteins after the control for gene essentiality (Julenius and Pedersen 2006), suggesting that extracellularity does not directly influence protein evolutionary rate. Here, we show that this is not the case in mammals. More importantly, the impact of extracellularity on mammalian protein evolutionary rate is not only independent from that of gene essentiality but also the greatest among all the factors examined.
To investigate the influence of extracellularity on the evolutionary rate of mammalian proteins, we chose mouse (Mus musculus) as our focal species for the comprehensiveness of its genomic (Waterston et al. 2002) and transcriptomic (Su et al. 2004) data. Based on the “cellular component” terms in Gene Ontology (GO; www.geneontology.org), we treated proteins exclusively located outside and inside the cell membrane as extracellular and intracellular proteins, respectively (see supplementary fig S1, Supplementary Material online and Materials and Methods). Following our previous work (Liao et al. 2006), we defined mouse essential genes by knockout phenotypes of premature death or sterility (see Materials and Method). To study the impact of a factor on the protein evolutionary rate, one should compare closely rated species (Zhang and He 2005) because gene properties (e.g., subcellular localization and essentiality; Liao and Zhang 2008; Qian and Zhang 2009) and evolutionary rates may change in evolution. We thus used one-to-one orthologs between mouse and rat (Rattus norvegicus) to estimate the rates of synonymous (dS) and nonsynonymous (dN) substitutions. Secreted proteins often contain a rapidly evolving signal peptide (Williams et al. 2000). To avoid overestimating substitution rates of extracellular proteins, signal peptides were removed prior to estimating dN and dS. We found that extracellular proteins are enriched with proteins related to immune response (P = 2.54e−28, χ2 test). Because many immune-related proteins are subject to positive selection (Hughes 1999) and because X-linked genes tend to be fast evolving (Vicoso and Charlesworth 2006), these proteins were excluded. Our final data set for subsequent analysis included 3,069 mouse–rat orthologs with information about gene essentiality and subcellular localization in mouse. Among them, 1,740 are intracellular and 288 are extracellular (see supplementary fig. S1, Supplementary Material online).
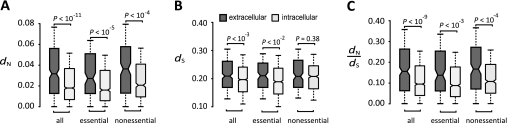
We found that extracellular proteins have an average mouse–rat dN (0.047) 61% higher than that of intracellular proteins (0.029; P = 7.1E-12, Mann–Whitney U test; fig. 1A). Because the average dS for extracellular (0.24) and intracellular (0.21) proteins differ by only 13.5% (fig. 1B), elevated mutation rate does not fully explain the difference in dN. Significantly higher dN/dS of extracellular proteins (average = 0.202) than intracellular proteins (0.132; fig. 1C) suggests that extracellular proteins are subject to more frequent or stronger positive selection or relaxed purifying selection. Mammalian essential proteins tend to evolve slowly (Liao et al. 2006). We found that intracellular proteins contain a higher proportion of essential genes (1104/1740 = 63.5%) than extracellular proteins (125/288 = 43.4%; P = 1.13e−10, χ2 test). However, dN and dN/dS of extracellular proteins are still significantly greater than intracellular proteins (fig. 1A and C), even when only essential or only nonessential genes are considered. Clearly, extracellularity impacts the rate of mammalian protein evolution independently from gene essentiality.
FIG. 1.
Comparison of (A) dN, (B) dS, and (C) dN/dS between extracellular and intracellular proteins. The upper quartile, median, and lower quartile are indicated in each box; the bars indicate semiquartile ranges. The P values (U test) for the hypothesis of no difference between extracellular and intracellular proteins are shown above each panel.
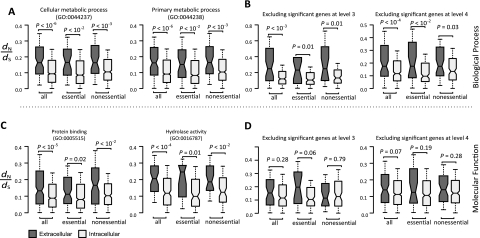
In addition to essentiality and extracellularity, determinants of the rate of mammalian protein evolution also include expression level, tissue-specificity, number of interacting proteins, and gene compactness (i.e., length of introns and untranslated regions, or UTRs; Liao et al. 2006; Liang and Li 2007). We compared the relative importance of these factors by performing Spearman's rank correlation between dN and each of the above factors. Important factors are expected to show stronger rank correlations with dN (Xia et al. 2009). We found that extracellularity is the most important factor in determining dN and dN/dS among all the factors examined (table 1). Furthermore, the correlation between extracellularity and dN is not substantially reduced after the control of other factors (table 2). Partial correlation analysis may have limitations under certain conditions (Drummond et al. 2006; Kim and Yi 2007). We thus conducted a principle component regression analysis of the same data. Consistent with the results from the partial correlation analysis (tables 1 and 2), we found that extracellularity contributes most to the first principle component that explains the variance in mouse–rat dN, dS, and dN/dS (supplementary table S1, Supplementary Material online). It is possible that the evolutionary rate difference between intracellular and extracellular proteins is a by-product of different distributions of GO terms among the two groups of proteins. However, higher dN/dS for extracellular proteins than intracellular proteins was observed even when we compared proteins of the same GO terms (fig 2A and C) or after excluding genes with differentially distributed GO terms (fig. 2B and D). Twenty-three GO categories are significantly differently distributed between intracellular and extracellular proteins and each contain at least 25 essential extracellular proteins, 25 nonessential extracellular proteins, 25 essential intracellular proteins, and 25 nonessential intracellular proteins (supplementary table S2, Supplementary Material online). With the exception of two GO categories, median dN/dS of extracellular proteins are significantly higher than that of intracellular proteins when proteins of the same essentiality are compared within each of the GO categories (supplementary table S2, Supplementary Material online). We also repeated our analysis by removing proteins of unknown molecular functions, proteins not involved in any known biological process, and proteins located in synapse and obtained essentially the same results (supplementary figs. S3 and S4, Supplementary Material online). Together, these results indicate that extracellularity has a major, and likely direct, impact on mammalian protein evolution.
Table 1.
Rank Correlations of Various Factors with dN or dN/dS
| Gene properties | ρ (P value) for correlation with dN | ρ (P value) for correlation with dN/dS |
| Mammals | ||
| Extracell | 0.177 (6.28e−11) | 0.166 (1.17e−09) |
| 5′UTR | −0.144 (9.75e−08) | −0.133 (9.71e−07) |
| Essen | −0.128 (2.21e−06) | −0.101 (1.88e−4) |
| TissSpcf | 0.122 (6.88e−06) | 0.096 (4.23e−4) |
| KPPI | −0.104 (1.21e−4) | −0.103 (1.42e−4) |
| 3′UTR | −0.085 (1.71e−3) | −0.079 (3.71e−3) |
| Intron | −0.075 (5.72e−3) | −0.077 (4.48e−3) |
| ExpLev | −0.038 (0.165) | −0.060 (0.028) |
| Yeasts | ||
| ExpLev | −0.541 (2.65e−215) | −0.473 (1.22e−158) |
| Essen | 0.197 (2.73e−26) | 0.202 (1.57e−27) |
| KPPI | −0.122 (5.80e−11) | −0.151 (7.36e−16) |
| Extracell | 0.022 (0.246) | 0.022 (0.232) |
NOTE.—Extracell is 1 for extracellular proteins and 0 for intracellular proteins. Essen is 1 for essential genes and 0 for nonessential genes. “UTR” is UTR length and “Intron” is average length per intron. “KPPI” is the number of interacting proteins. “TissSpcf” is tissue-specificity. “ExpLev” is gene expression level. P values show the probabilities of the observations under the hypothesis of no correlation. The analysis is based on 1,350 mouse–rat orthologs or 2,840 Saccharomyces cerevisiae–S. paradoxus orthologs.
Table 2.
Partial Rank Correlations of Various Factors with dN or dN/dS
| Extracell | controlled property | ρ (P value) for correlation with dN | ρ (P value) for correlation with dN/dS |
| Mammals | ||
| Extracell | Intron | 0.175 (6.50e−11) | 0.163 (1.30e−09) |
| Extracell | ExpLev | 0.174 (9.55e−11) | 0.159 (3.16e−09) |
| Extracell | 3′UTR | 0.172 (1.63e−10) | 0.160 (2.79e−09) |
| Extracell | KPPI | 0.166 (6.21e−10) | 0.154 (1.04e−08) |
| Extracell | 5′UTR | 0.166 (7.03e−10) | 0.154 (9.99e−09) |
| Extracell | Essen | 0.160 (2.96e−09) | 0.151 (1.98e−08) |
| Extracell | TissSpcf | 0.158 (4.67e−09) | 0.150 (2.63e−08) |
| Yeasts | ||
| Extracell | ExpLev | 0.043 (0.023) | 0.040 (0.035) |
| Extracell | KPPI | 0.015 (0.415) | 0.015 (0.439) |
| Extracell | Essen | 0.011 (0.562) | 0.011 (0.547) |
NOTE.—See note of table 1 for Extracell, Essen, UTR, Intron, KPPI, TissSpcf, and ExpLev. The factor before “|” is the factor being examined and that after “|” is the factor being controlled for. P values show the probabilities of the observations under the hypothesis of no correlation. The analysis is based on 1,350 mouse–rat orthologs or 2,840 Saccharomyces cerevisiae–S. paradoxus orthologs.
FIG. 2.
Higher dN/dS of extracellular proteins than intracellular proteins are observed even after the control of biological processes (A, B) or molecular functions (C, D). (A) and (C) were performed by comparing proteins of the same GOs at level 3 (left and right are top first and second differentially enriched terms, respectively), whereas (B) and (D) were performed by removing all genes with significantly differentially enriched GO terms at level 3 (left) or level 4 (right). See legend of figure 1 for details of the boxplot and supplementary figure S2 (Supplementary Material online) for the results of dN and dS.
The influence of extracellularity on protein evolutionary rate differs greatly between what we found in mammals and what was reported in yeasts (Julenius and Pedersen 2006). To examine whether the difference is due to the different analytical approaches used, we applied the same analytical procedures to the orthologs of yeast species Saccharomyces cerevisiae and S. paradoxus (see Materials and Methods). Our results for yeasts are consistent with those previously published (table 1). That is, extracellularity has no effect on yeast protein evolutionary rate after the control of gene essentiality (Julenius and Pedersen 2006), and expression level is the most important rate determinant in yeast (Drummond et al. 2006). It should be noted that, compared with what was reported previously (Drummond and Wilke 2008), we observed a weaker correlation between expression level and dN for mammalian proteins (tables 1 and 2). This difference is probably due to the smaller number of genes used here, as there are fewer genes with all the information needed in our partial correlation analysis. Although the reduction in sample size may have resulted in weaker correlations, it should not have changed the relative importance of different factors as shown in tables 1 and 2.
The different impacts of extracellularity on protein evolutionary rates in yeasts and mammals can potentially be explained in two ways. First, extracellularity has qualitatively different meanings in these species because, for yeasts, secreted proteins are outside the organisms, whereas for mammals, they are largely inside the organisms. However, this difference implies that properties of extracellular and intracellular proteins should be more similar in mammals than in yeasts, which does not explain our observation on the rate of protein evolution. Second, secreted proteins involved in the biological processes that are present in mammals but not in yeasts evolve rapidly. However, figure 2 and supplementary table S2 (Supplementary Material online) showed that, even within the same functional categories, extracellular proteins evolve faster than intracellular proteins, suggesting that the faster evolution of extracellular proteins is not attributable to special functions of these proteins. Because only a small fraction of mammalian genes are subject to recurrent positive selection and genes most likely to be subject to such selection (i.e., immunity genes) have been removed from our analysis, the observed evolutionary rate difference between extracellular and intracellular proteins is most likely owing to differed purifying selection acting on them. But, the exact biological factors that cause the difference in purifying selection remain to be explored.
Materials and Methods
The annotations, sequences, and orthologous relationships of mouse and rat genes were retrieved from Ensembl version 53 (www.ensembl.org), whereas those of yeast genes were obtained from the Saccharomyces Genome Database (www.yeastgenome.org). Based on GO, immune-related proteins have the annotation of GO:0002376 (immune system process). Extracellularity was defined by the GO terms for cellular component (see supplementary fig. S1, Supplementary Material online). Here, a GO term includes all its child GO terms. Essentiality mouse genes were defined based on Mouse Genome Informatics 4.21 (www.informatics.jax.org), following Liao and Zhang (2007). Essentialities and expression levels of yeast genes were obtained from Zhang and He (2005). A zero fitness upon gene deletion is used to define essential genes in both the yeast and mouse. Properties of mouse gene expression were defined based on the microarray data of 61 mouse tissues (Su et al. 2004). Expression level was calculated by averaging expression signals in the 61 tissues, whereas tissue-specificity (τ), which ranges from 0 to 1 (higher values indicate stronger tissue-specificity), was calculated according to Liao et al. (2006). Experimentally verified yeast protein–protein interaction (PPI) data were obtained from Batada et al. (2007), and those for human were compiled from six sources: Human Protein Reference Database (www.hprd.org), Munich Information center for Protein Sequences (mips.helmholtz-muenchen.de), Molecular INTeraction database (mint.bio.uniroma2.it), Reactome (www.reactome.org), IntAct (www.ebi.ac.uk), and Database of Interacting Proteins (dip.doe-mbi.ucla.edu). The human ortholog's number of interacting proteins (KPPI) was used as a proxy for a mouse protein's KPPI.
To calculate mammalian or yeast protein evolutionary rate, signal peptides, annotated by SPdb (proline.bic.nus.edu.sg/spdb/), were removed. Orthologous coding sequences without signal peptides were aligned following the protein alignment by ClustalW (www.ebi.ac.uk/clustalw/). When a gene has multiple isoforms, the longest isoform was used. Values of dN and dS between mouse and rat and between S. cerevisiae and S. paradoxus were computed using PAML 4 (Yang 2007).
Supplementary Material
Supplementary figures S1–S4 and supplementary tables 1 and 2 are available at Genome Biology and Evolution online (http://www.oxfordjournals.org/our_journals/gbe/).
Acknowledgments
This project was supported by Taiwan National Health Research Institutes intramural funding to B.-Y.L. and US National Institutes of Health research grants to J.Z.
References
- Batada NN, et al. Still stratus not altocumulus: further evidence against the date/party hub distinction. PLoS Biol. 2007;5:e154. doi: 10.1371/journal.pbio.0050154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Good JM, Nachman MW. Adaptive evolution of proteins secreted during sperm maturation: an analysis of the mouse epididymal transcriptome. Mol Biol Evol. 2008;25:383–392. doi: 10.1093/molbev/msm265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Raval A, Wilke CO. A single determinant dominates the rate of yeast protein evolution. Mol Biol Evol. 2006;23:327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002;296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Adaptive evolution of genes and genomes. New York: Oxford University Press; 1999. [Google Scholar]
- Julenius K, Pedersen AG. Protein evolution is faster outside the cell. Mol Biol Evol. 2006;23:2039–2048. doi: 10.1093/molbev/msl081. [DOI] [PubMed] [Google Scholar]
- Kim SH, Yi SV. Understanding relationship between sequence and functional evolution in yeast proteins. Genetica. 2007;131:151–156. doi: 10.1007/s10709-006-9125-2. [DOI] [PubMed] [Google Scholar]
- Liang H, Li WH. Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends Genet. 2007;23:375–378. doi: 10.1016/j.tig.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Liao BY, Scott NM, Zhang J. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Mol Biol Evol. 2006;23:2072–2080. doi: 10.1093/molbev/msl076. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Mouse duplicate genes are as essential as singletons. Trends Genet. 2007;23:378–381. doi: 10.1016/j.tig.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci U S A. 2008;105:6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino T, Gojobori T. The evolutionary rate of a protein is influenced by features of the interacting partners. Mol Biol Evol. 2006;23:784–789. doi: 10.1093/molbev/msj090. [DOI] [PubMed] [Google Scholar]
- Qian W, Zhang J. Protein subcellular relocalization in the evolution of yeast singleton and duplicate genes. Genome Biol Evol. 2009;2009:198–204. doi: 10.1093/gbe/evp021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics. 2004;168:373–381. doi: 10.1534/genetics.104.028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Williams EJ, Pal C, Hurst LD. The molecular evolution of signal peptides. Gene. 2000;253:313–322. doi: 10.1016/s0378-1119(00)00233-x. [DOI] [PubMed] [Google Scholar]
- Winter EE, Goodstadt L, Ponting CP. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Franzosa EA, Gerstein MB. Integrated assessment of genomic correlates of protein evolutionary rate. PLoS Comput Biol. 2009;5:e1000413. doi: 10.1371/journal.pcbi.1000413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang J, He X. Significant impact of protein dispensability on the instantaneous rate of protein evolution. Mol Biol Evol. 2005;22:1147–1155. doi: 10.1093/molbev/msi101. [DOI] [PubMed] [Google Scholar]