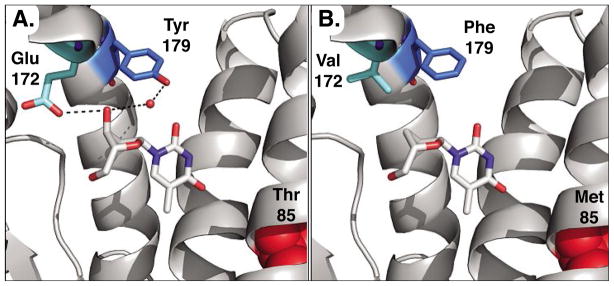
Fig. 7.
Structure model of active sites in DmdNK (A) and R4.V3 (B). The location of the three relevant mutations Glu172Val, Tyr179Phe, and Thr85Met are highlighted in DmdNK with thymidine, bound in the phosphoryl acceptor site (PDB code: 1OT3).[43] A) Glu172 in the wild-type enzyme forms a direct hydrogen-bonding interaction with the 3′-hydroxyl group of the substrate while Tyr179 contacts the same position via hydrogen bonding to a water molecule (red sphere) in the active site. B) Directed evolution of DmdNK for ddT phosphorylation introduces two hydrophobic residues in position 172 and 179, better accommodating the binding of the nucleoside analog. Separately, the mutation of Thr85 to Met is believed to slightly enlarge the binding pocket for the nucleobase portion by inducing a conformational shift of the helical region.