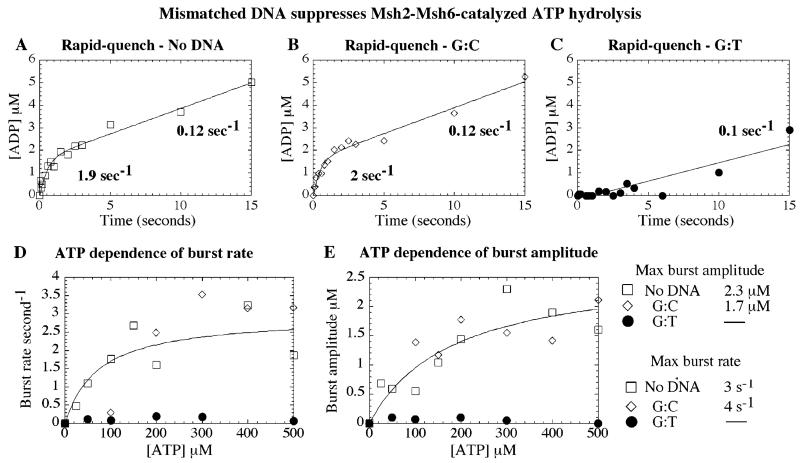
Figure 5.
Mismatched DNA binding to Msh2-Msh6 suppresses ATP hydrolysis. Pre-steady-state acid quench assays were performed at 20 °C with 2 μM Msh2-Msh6 and 0–500 μM α32P-ATP, ± 3 μM DNA. (A) A representative ATPase time course in the absence of DNA and with 500 μM ATP shows a burst of ATP hydrolysis, with burst rate 1.9 s−1, amplitude 1.6 μM, and turnover rate 0.12 s−1. (B) The ATPase kinetics are nearly identical with G:C DNA. (C) With G:T DNA, however, only a linear 0.1 s−1 rate is observed. (D) ATP dependence of the kinetic parameters fit to a hyperbola yields a maximum burst rate of 3 and 4 s−1 and apparent Kd = 73 and 100 μM and (E) maximum burst amplitude of 2.3 and 1.7 μM and apparent Kd = 160 and 70 μM for Msh2-Msh6 alone or in the presence of G:C DNA, respectively (fit shown only for no DNA condition, for clarity).