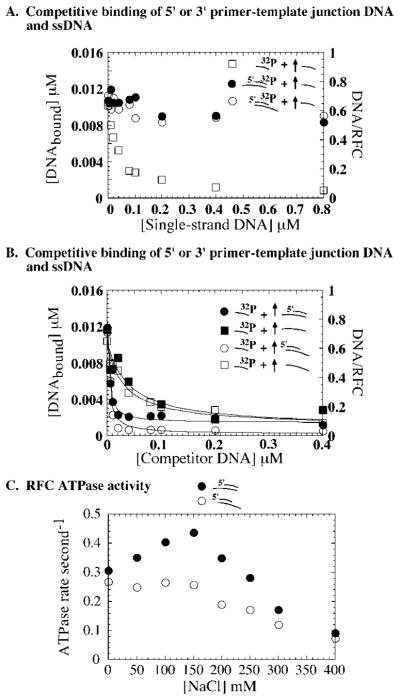
Fig. 7. A 5′ or 3′ primer-template junction supports specific interaction between RFC and primed DNA.
A, binding of 0.02 μM 32P-ssDNA (□), 32P-31/56 5′ primer junction DNA (●), or 32P-31/56 3′ primer junction DNA (○) to RFC (0.016 μM) in the presence of 0–1 μM unlabeled ssDNA competitor. B, a complementary experiment in which 32P-ssDNA 0.02 μM binding to RFC (0.016 μM) is competed with 0–0.4 μM unlabeled 31/56 5′ primer junction DNA (●), 56-mer template ssDNA (■), 31/56 3′ primer junction DNA (○), and the corresponding 56-mer template ssDNA (□). For both 56-mer single-stranded DNAs, the K1/2 is 0.035 μM, indicating simple competition between the 31- and 56-mer ssDNAs. In contrast, both 3′ and 5′ primer junction DNA competitors yield a K1/2 of 0.004 μM. C, the ATPase activity of RFC (0.2 μM) measured in the presence of 5′ primer junction DNA (●) or 3′ primer junction DNA (○), as described under “Experimental Procedures.” The peak steady-state ATPase rate is 0.45 s−1 for the 5′ primer junction (similar to full primer-template DNA) and 0.25 s−1 for 3′ primer junction (similar to ssDNA).