Abstract
Prokaryotic MutS and eukaryotic Msh proteins recognize base pair mismatches and insertions or deletions in DNA and initiate mismatch repair. These proteins function as dimers (and perhaps higher order oligomers) and possess an ATPase activity that is essential for DNA repair. Previous studies of Escherichia coli MutS and eukaryotic Msh2–Msh6 proteins have revealed asymmetry within the dimer with respect to both DNA binding and ATPase activities. We have found the Thermus aquaticus MutS protein amenable to detailed investigation of the nature and role of this asymmetry. Here, we show that (a) in a MutS dimer one subunit (S1) binds nucleotide with high affinity and the other (S2) with 10-fold weaker affinity, (b) S1 hydrolyzes ATP rapidly while S2 hydrolyzes ATP at a 30–50-fold slower rate, (c) mismatched DNA binding to MutS inhibits ATP hydrolysis at S1 but slow hydrolysis continues at S2, and (d) interaction between mismatched DNA and MutS is weakened when both subunits are occupied by ATP but remains stable when S1 is occupied by ATP and S2 by ADP. These results reveal key MutS species in the ATPase pathway; S1ADP–S2ATP is formed preferentially in the absence of DNA or in the presence of fully matched DNA, while S1ATP–S2ATP and S1ATP–S2ADP are formed preferentially in the presence of mismatched DNA. These MutS species exhibit differences in interaction with mismatched DNA that are likely important for the mechanism of MutS action in DNA repair.
Mismatch repair maintains genomic integrity by correcting mispaired bases and insertions or deletions that occur in DNA due to errors in DNA replication or recombination. The process initiates with a mismatch recognition phase, in which MutS protein binds to the distortion in the DNA duplex, followed by excision of the offending DNA strand, catalyzed by helicase and exonuclease enzymes, and finally DNA resynthesis and ligation of the new strand, apparently by the normal DNA replication machinery (1). In Escherichia coli, after MutS (a homodimer) binds the mismatch, it interacts with MutL (also a homodimer), resulting in stimulation of MutH endonuclease activity and nicking of the mismatch-containing DNA strand to initiate strand excision (2–4). Since these core components of the DNA mismatch repair system were identified in E. coli, homologues of MutS and MutL have been discovered and analyzed in numerous other organisms, including humans. Eukaryotic MutS proteins function as heterodimers, such as Msh2–Msh6,1 which recognizes base pair mismatches and small insertion or deletion loops, and Msh2–Msh3, which appears to be specific for insertion or deletion loops. MutL homologues, such as Mlh1–Pms2 heterodimers, interact with the Msh proteins as well as single-stranded DNA and appear to couple mismatch recognition to initiation of excision and DNA repair, as observed in E. coli; their exact function and mechanism of action is not entirely known, especially because no eukaryotic homologues of MutH have been found and it is not clear exactly how strand-specific excision is initiated (5–9).
All MutS proteins identified thus far also possess an ATPase activity that is essential for DNA repair. These proteins belong to the ATP binding cassette (ABC) family of ATPases and have Walker A (ATP-binding P loop) and Walker B (Mg2+-binding) motifs that catalyze ATP binding and hydrolysis (10–15). How MutS proteins utilize ATP for DNA repair is a question of great interest to researchers trying to understand the workings of the mismatch repair system. The most well examined—and the most debated—role of MutS ATPase activity is in the coupling of DNA mismatch recognition to initiation of strand excision. MutS proteins undergo changes in conformation in the presence of ATP and its nonhydrolyzable analogue, ATPγS (16–20), and MutS affinity for mismatched DNA is lowered in the presence of ATP or ATPγS, which manifests as the protein sliding off the ends of linear DNA substrates if they are unblocked (17, 21, 22). This evidence suggests that following ATP binding, MutS moves away from the site of the mismatch, perhaps using a translocation-based mechanism to communicate with downstream repair proteins, search for the excision site, or both; MutS ATPase activity is thought to facilitate its movement on DNA either by repeated cycles of ATP binding and hydrolysis (22, 23) or perhaps by switching the protein into an ATP-bound clamp-like form that can slide on the duplex (17, 24, 25). On the other hand, there is also evidence to indicate that MutS•MutL complexes form at the mismatch site and direct excision at distant sites via a DNA looping mechanism (13, 26–28); in this model, MutS ATPase activity is thought to facilitate mismatch verification by triggering differential binding of MutS to mismatched versus fully matched DNA and facilitate interactions with downstream repair proteins as well.
Another property of MutS proteins, recognized only in the past few years, further complicates analysis of its mechanism of action in DNA repair—subunits of the MutS dimer exhibit asymmetry in both their DNA binding and ATPase activities. The crystal structure of E. coli MutS dimer shows one Walker A site occupied by ADP, while the other site remains nucleotide-free (15). Nucleotide-binding analyses of E. coli MutS (29, 30) and Saccharomyces cerevisiae Msh2–Msh6 (31) proteins revealed differential affinities of the two subunits for ATP, ADP, and nonhydrolyzable ATP analogues. E. coli MutS dimer was also shown to be capable of binding a nucleotide di- and triphosphate simultaneously (29). These results predicted asymmetry in the ATP hydrolysis activity of the two subunits, raising the possibility of up to nine different nucleotide-bound and nucleotide-free species occurring in the ATPase reaction. Studies of S. cerevisiae and human Msh2–Msh6 wild-type and mutant ATPase mixed heterodimers appear to support this hypothesis, becasue they show differential effects of mutations in Msh2 versus Msh6 on the steady-state ATPase activity (18, 32, 33); these results cannot be considered unequivocal evidence for asymmetric ATP hydrolysis, because the steady-state rate does not reflect ATP hydrolysis but rather a slow, rate-limiting step occurring in the reaction after ATP hydrolysis (31, 34). The S. cerevisiae Msh2–Msh6 study provided more convincing evidence of asymmetric hydrolytic activity, because it showed only one ATP molecule hydrolyzed rapidly per dimer per turnover; although it remained possible that the observed half-site reactivity reflected a mixed population of active and inactive or partially active proteins in the reaction (31).
The asymmetry in ATPase activity within the MutS dimer coincides with asymmetry in its DNA binding activity. In both Thermus aquaticus and E. coli MutS•DNA crystal structures, only one subunit in the MutS dimer inserts a phenylalanine residue into DNA that stacks against the mismatched/bulged base; other hydrogen-bonding and van der Waals contacts between the two subunits and DNA are also asymmetric (14, 15). In the eukaryotic heterodimers, Msh6 possesses an equivalent phenylalanine, but Msh2 does not (35, 36). Interestingly, in the E. coli MutS structure, phenylalanine from the ADP-bound subunit is inserted into the mismatch (15). Also, mismatched DNA binding to S. cerevisiae Msh2–Msh6 inhibits the rapid hydrolysis of one ATP molecule that occurs in the absence of DNA (31). These results imply that asymmetric ATPase activity of the MutS dimer is linked to its interactions with DNA during mismatch repair.
We have attempted to clarify this link by measuring the ATP binding and hydrolysis activity of each subunit in the T. aquaticus MutS dimer to identify the nucleotide-bound MutS species formed in the reaction and by measuring the interaction of these species with mismatched DNA. The results reveal dynamic MutS–DNA interactions during the course of the ATPase reaction that may be important for mismatch recognition, for signaling initiation of excision and DNA repair, or for both.
EXPERIMENTAL PROCEDURES
DNA, Nucleotides, and Other Reagents
Synthetic DNAs were purchased from Integrated DNA Technologies and purified by denaturing polyacrylamide gel electrophoresis. The mismatched duplex substrate was prepared by annealing 22-template, 5′ GGA CGA GCC GCC GCT AGC GTC G 3′, and 23+T complement, 5′ GCG ACG CTA GCG TGC GGC TCG TC 3′. The matched duplex substrate was prepared by annealing 23-template, 5′ GGA CGA GCC GCT CGC TAG CGT CG 3′, and A:T complement, 5′ GCG ACG CTA GCG AGC GGC TCG TC 3′. The DNAs were mixed in 1:1 ratio (20–100 µM concentration in 20 mM Tris-HCl, pH 8, 75 mM NaCl) and heated to 95 °C followed by slow cooling over 6–8 h to 25 °C (nondenaturing PAGE analysis of the products revealed >95% duplex DNA). 32P-labeled DNA was prepared as described previously (31). Radioactive nucleotides, α32P-ATP, γ32P-ATP, and 35S-ATPγS, were purchased from Perkin-Elmer Life Sciences. α32P-ADP was prepared as described previously (31). Nonradioactive nucleotides were purchased from Sigma Aldrich. PEI cellulose-F TLC plates were purchased from EM Science and nitrocellulose membranes from Schleicher and Schuell.
Protein Purification
T. aquaticus MutS was overexpressed and purified as described previously (37) with modifications; overexpression was carried out in E. coli BL21(DE3) cells (Novagen), and the protein was purified by ion-exchange chromatography over a 25 mL Q-Sepharose column (Amersham) using a 50–350 mM NaCl gradient. Phosphate binding protein (PBP) was purified and labeled with MDCC as described previously (38, 39).
Nucleotide Binding Assays
ATP, ATPγS, and ADP binding to MutS were measured by nitrocellulose membrane filtration as described previously (31). Briefly, 15 µL reactions containing 1 µM MutS and 0–200 µM ATP and 0.3 µCi of α32P-ATP (3 min at 4 or 25 °C), 0–200 µM ADP and 0.3 µCi of α32P-ADP (10 min at 25 °C), or 0–200 µM ATPγS and 0.3 µCi of 35S-ATPγS (15 min at 25 °C) were incubated in nucleotide binding buffer (50 mM Hepes, pH 7.8, 5 mM MgCl2, 150 mM KCl, and 5% glycerol). Ten microliter aliquots of the reaction were filtered through the membrane. The membranes were washed before and after filtration with 150 µL of wash buffer (50 mM Hepes, pH 7.8, 5 mM MgCl2). One microliter aliquots were spotted onto a separate membrane to measure total nucleotide in the reaction. The molar amount of nucleotide bound to MutS dimer was determined and plotted versus nucleotide concentration. The binding isotherms were fit to an equation describing 1:1 protein–ligand interaction for ATP binding, [N•M] = 0.5{(Kd + [Nt] + [Mt]) − [(Kd + [Nt] + [Mt])2 − 4[Nt][Mt]]1/2}, where N•M is the molar amount of ATP bound to MutS, Nt and Mt are total ATP and MutS concentrations, respectively, and Kd is the apparent dissociation constant, or to an equation describing 1:2 protein–ligand interaction for ADP and ATPγS binding, [N•M]/[Mt] = K1[Nf] + 2K1K2−[Nf]2/(1 + K1[Nf] + 2K1K2[Nf]2), where N•M is the fraction of nucleotide bound to MutS, Mt is total MutS concentration, Nf is free nucleotide concentration, and K1 and K2 are apparent stepwise association constants.
The mixed-nucleotide experiments were performed similarly with 5 µM unlabeled ATPγS incubated first with 1 µM MutS for 10 min, followed by addition of 0–150 µM ADP and 0.3 µCi of 32P-ADP and further incubation for 3 min before filtration through the membrane. The complementary experiment measuring ATPγS bound to MutS was performed in an identical manner with 5 µM ATPγS and 0.3 µCi of 35S-ATPγS and 0–150 µM unlabeled ADP in the reaction.
The rate of dissociation of ATPγS from MutS was measured by incubating 1 µM MutS with 12 µM ATPγS and 0.3 µCi of 35S-ATPγS at 25 °C for 15 min, followed by addition of 500 µM ATP or ADP (and 5 mM Mg2+) chase and filtration through the membrane at times ranging from 20 s to 5 min. Decay of the protein–nucleotide complex over time was fit to a single-exponential equation.
The MutS preparation was tested for possible ATP and ADP contamination by an ATP-dependent bioluminescent assay (Sigma Aldrich) as described previously for S. cerevisiae (31) and human Msh2–Msh6 (22).
ATPase Assays
Temperature dependence of MutS ATPase activity was measured at 25–80 °C with 1 µM MutS and 1 mM α32P-ATP in nucleotide binding buffer in the absence or presence of 2 µM DNA. ATP hydrolysis was quantitated by PEI cellulose thin-layer chromatography, and kcat values were determined as described previously (31).
Pre-steady-state ATPase assays were performed on a KinTek Corp. quench-flow instrument (Austin, TX) with 6 µM MutS dimer and 1 mM α32P-ATP at various temperatures (25–70 °C) with or without 10 µM DNA in nucleotide binding buffer. Sixteen microliters of MutS (with or without DNA) was mixed rapidly with 16 µL of ATP and quenched after varying times (0.01–10 s) with 35 µL of 0.7 M formic acid (final concentrations, 3 µM MutS dimer, 500 µM 32P-ATP, and 5 µM DNA). The molar amount of ADP formed was plotted versus time and fit to [ADP] = A(1 − e−kt) + Vt, where A and k are burst amplitude and rate constant, respectively, V is the velocity of the linear phase, and t is reaction time, for experiments with no DNA or matched DNA. Data with mismatched DNA were fit to a linear equation. Experiments containing ATPγS were performed similarly, except 6 µM MutS was preincubated with 10 µM ATPγS in the absence or presence of 10 µM mismatched DNA.
Pulse–chase experiments were performed similarly, except 35 µL of 10 mM unlabeled Mg2+ ATP chase (final concentration) was added to the reaction after varying times (0.004–2.5 s). After chase time equivalent to about five turnovers (20 s; kcat = 0.3 s−1), the reactions were quenched with 70 µL of 0.7 M formic acid and analyzed as above.
Chase-time experiments were performed by mixing 3 µM MutS with 500 µM 32P-ATP (final concentrations) for 20 ms prior to addition of 10 mM unlabeled Mg2+ ATP chase (final concentrations) and quenching the reaction after varying times (0.1–20 s) with 35 µL of 0.7 M formic acid.
Stopped-flow kinetic experiments were performed on a Kintek SF-2001 stopped-flow instrument (KinTek Corp., Austin, TX) to measure the rates of ATP hydrolysis and phosphate release catalyzed by MutS. Phosphate (Pi) release was assayed in real-time using a fluorescent probe (MDCC) attached to E. coli PBP as described previously (39–41). Change in fluorescence of MDCC–PBP upon Pi binding was monitored using an excitation wavelength of 425 nm and emission above 450 nm (cutoff filter, Corion LL-450 F). A coupled enzyme reaction (Mop) containing 200 µM 7-methyl guanosine and 0.01 U/mL purine nucleoside phosphorylase was used in all reactions to sequester contaminant Pi as ribose-1-phosphate, because MDCC–PBP is sensitive to micromolar concentrations of Pi. A Pi calibration curve relating the PBP–MDCC fluorescence signal to Pi concentration was generated prior to each experiment, as described previously (39). All reactions and syringes were “mopped” for at least 45 min before each experiment.
Pi release experiments were carried out at 40 °C in buffer containing 20 mM Tris-HCl, pH 7.7, 5 mM MgCl2, and Mop. A 60 µL solution of 2 µM MutS incubated with 8 µM MDCC–PBP in the absence or presence of 4 µM DNA was mixed rapidly with an equal volume of ATP (10–2000 µM), and the change in fluorescence was monitored over time in the observation cell (final concentrations, 1 µM MutS, 5–1000 µM ATP, 2 µM DNA, 4 µM MDCC–PBP). Background fluorescence, measured for each reaction by omitting Mg2+ to prevent ATP hydrolysis, was subtracted from the traces. Raw data obtained by averaging at least five traces were divided by the slope from the Pi calibration curve to determine the molar amount of Pi released during the reaction. Data were fit to a burst equation or a linear equation as described above. Experiments containing ATPγS were performed in a similar manner, except 1–200 µM ATPγS was preincubated with 2 µM MutS in the absence or presence of 4 µM DNA prior to rapid mixing with ATP.
DNA Binding Assays
Nitrocellulose membrane filtration experiments were performed to measure protein–DNA interaction as described previously (31). Briefly, 15 µL reactions containing 1 µM MutS and 0–3 µM 32P-labeled DNA in DNA binding buffer (20 mM Hepes, pH 7.8, 5 mM MgCl2, 150 mM KCl, 0.1 mM EDTA, 0.1 mM DTT, and 5% glycerol) were incubated at 25 °C for 10 min in the absence or presence of ADP (250 µM) or ATPγS (10 or 250 µM), followed by membrane filtration. For the ATPγS and ADP experiment, MutS was preincubated with 10 µM ATPγS for 10 min followed by addition of 250 µM ADP and incubation for 3 min before membrane filtration. The membrane was washed before and after filtration with 125 µL of wash buffer (20 mM Hepes, pH 7.8, and 5 mM MgCl2). One microliter aliquots were spotted onto a separate membrane to measure total DNA in the reaction. Radioactivity on the membrane was quantitated on a PhosphorImager (Molecular Dynamics), and the molar amount of DNA bound to MutS was determined and plotted versus MutS concentration.
The rate of dissociation of DNA from MutS was measured by incubating 0.5 µM MutS dimer with 1 µM 32P-labeled +T DNA in DNA binding buffer for 10 min at 25 °C, followed by addition of 50 µM unlabeled +T DNA (and 5 mM Mg2+) chase (without nucleotides or mixed with 250 µM ATPγS or 500 µM ATP) and membrane filtration over time (0–500 s). The molar amount of DNA bound to MutS was quantitated as described above.
RESULTS
We report here an investigation of T. aquaticus MutS dimer aimed at clarifying the nature and consequences of asymmetry in the ATP-binding and -hydrolysis and DNA-binding activities of its subunits. We have employed rapid kinetic analysis of MutS ATPase activity and quantitative measurements of nucleotide and DNA binding to determine various nucleotide-bound forms of MutS dimer and examine their possible roles in the mismatch repair pathway. A key finding of this study is that following interaction with mismatched DNA, the MutS dimer appears to be stabilized in S1ATP–S2ATP and S1ATP–S2ADP forms, in contrast to the S1ADP–S2ATP form that is predominant in the absence of DNA or in the presence of fully matched DNA.
T. aquaticus MutS Dimer Binds Two Nucleotides with Differing Affinities
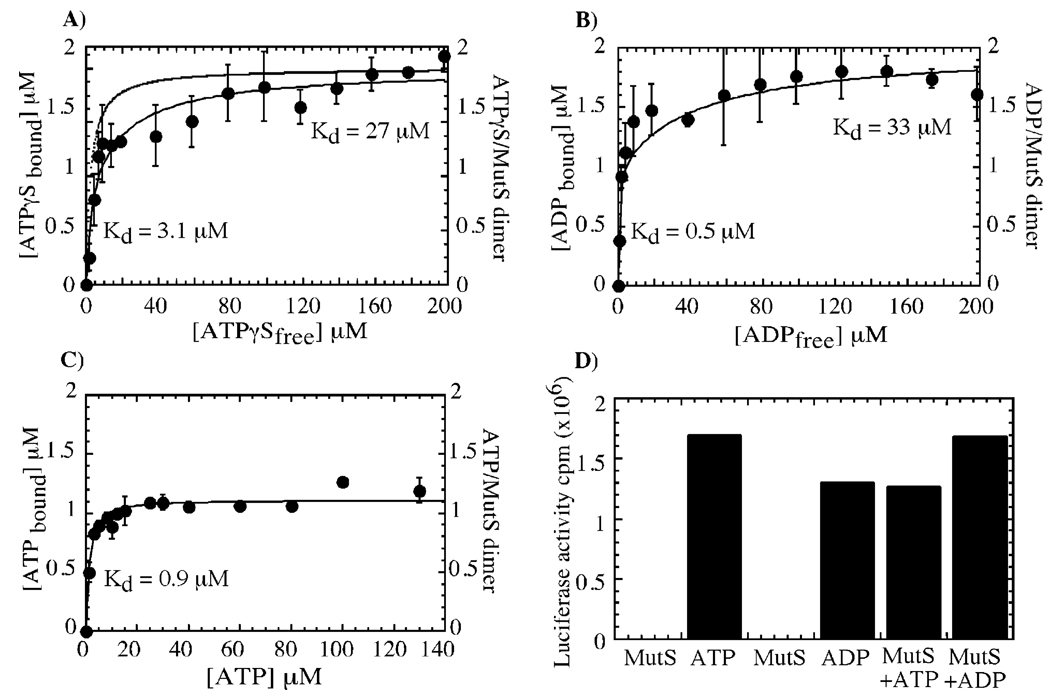
Nitrocellulose membrane filtration experiments were used to measure the stoichiometry and affinity of MutS binding to various nucleotides. Titration of 1 µM MutS dimer with increasing concentrations of the nonhydrolyzable ATP analogue 35S-ATPγS yielded a biphasic binding isotherm saturating at 2 µM ATPγS, that is, two ATPγS molecules bound per MutS dimer (Figure 1A; no ATPγS binding to the membrane was detectable in the absence of protein). MutS appears to bind one ATPγS with high affinity (Kd = 3.1 µM) and the other with about 10-fold lower affinity (Kd = 27 µM); note that the binding isotherm fit to a single equilibrium constant yields a Kd of 10 µM, which is incompatible with the 2.5 µM Kd measured for ATPγS binding to MutS in an in-solution assay under equilibrium conditions, Figure 3E. Accordingly, a simulated binding isotherm with the Kd set at 2.5 µM for both ATPγS molecules deviates significantly from the experimental data (Figure 1A). A similar experiment with 32P-ADP also showed two ADP molecules binding per MutS dimer with Kd values of 0.5 and 33 µM (Figure 1B). In contrast, the α32P-ATP binding isotherm saturates with only one ATP bound tightly to MutS (Kd = 0.9 µM, Figure 1C). Possible explanations for half-maximal binding observed with ATP and the differential binding affinities observed with ATPγS and ADP are that a large fraction of MutS already contains tightly bound nucleotide or that a large fraction of MutS is inactive or partially active. A luciferase-based bioluminescence assay that is sensitive to low nanomolar concentrations of ATP or ADP did not detect any nucleotide contamination in heat-denatured MutS (Figure 1D). We cannot directly rule out the possibility of two MutS populations in the reaction, about one-half capable of binding two nucleotides per dimer with high affinity (active) and the other binding two nucleotides per dimer with low affinity (inactive or partially active). However, our previous study of homologous S. cerevisiae Msh2–Msh6 protein also revealed two ATPγS molecules binding per dimer with Kd values of 4 and 17 µM (31). It seems unlikely that the similar nucleotide binding profiles exhibited by MutS proteins from two very different organisms are due to half inactive or partially active protein fractions in both preparations. Thus, we conclude that both subunits in the MutS homodimer are capable of binding nucleotides, although with differing affinities, and that we do not observe a second ATP binding to MutS perhaps because the interaction is too weak for the ATP to remain trapped on the membrane during the filtration and wash steps.
FIGURE 1.
T. aquaticus MutS dimer binds two nucleotides with differing affinities. In panel A, 35S-ATPγS binding to MutS (1 µM dimer) was measured by nitrocellulose membrane filtration. One ATPγS molecule binds MutS with high affinity (Kd = 3 ± 0.6 µM), and a second one binds with lower affinity (Kd = 27 ± 8 µM). In panel B, similarly, two ADP molecules bind MutS with differing affinities (Kd = 0.5 ± 0.05 and 33 ± 5 µM). In panel C, Only one ATP binding to MutS is detectable (Kd = 0.9 ± 0.2 µM). In panel D, bioluminescence assays performed with heat-denatured MutS indicate no tightly bound ATP or ADP contaminants in the preparation, as 100 nM MutS alone yields luciferase activity equivalent to 39 cpm compared with 1.69 × 106 and 1.33 × 106 cpm for 100 nM ATP and ADP standards, respectively. In control experiments, exogenous addition of 100 nM ATP or ADP to MutS yields luciferase activity equivalent to 1.35 × 106 and 1.68 × 106 cpm, respectively.
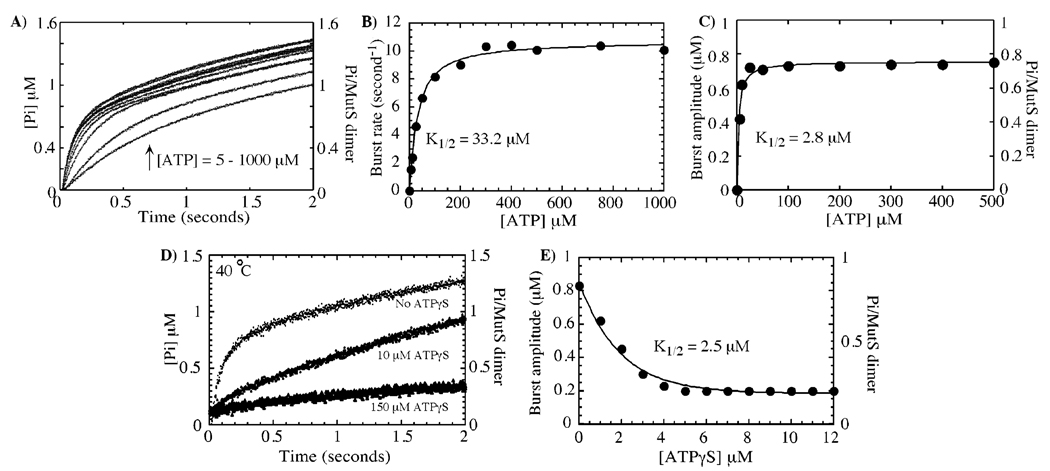
FIGURE 3.
The high-affinity ATP-binding site on MutS catalyzes rapid ATP hydrolysis. In panel A, phosphate-release kinetics were measured at increasing ATP concentrations and, in panel B, yielded a maximum burst rate constant of 10.7 ± 0.2 s−1 and K1/2 = 33.2 ± 2.5 µM, as well as, in panel C, a maximum amplitude of 0.8 ± 0.01 µM and K1/2 = 2.8 ± 0.4 µM for 1 µM MutS dimer at 40 °C. In panel D, preincubation of MutS (1 µM dimer) with increasing concentrations of ATPγS (0–200 µM) suppresses the rapid burst of hydrolysis (500 µM ATP in the reaction). In panel E, no burst phase can be detected at ATPγS concentrations higher than 5 µM; that is, following ATPγS binding to the high-affinity site (see Figure 1A), and a plot of decreasing burst amplitudes versus ATPγS concentration fit to a hyperbola yields K1/2 = 2.5 ± 0.7 µM (similar to the Kd for ATPγS binding to the high-affinity site on MutS, see Figure 1A).
Asymmetry in ATP Binding Is Recapitulated during ATP Hydrolysis
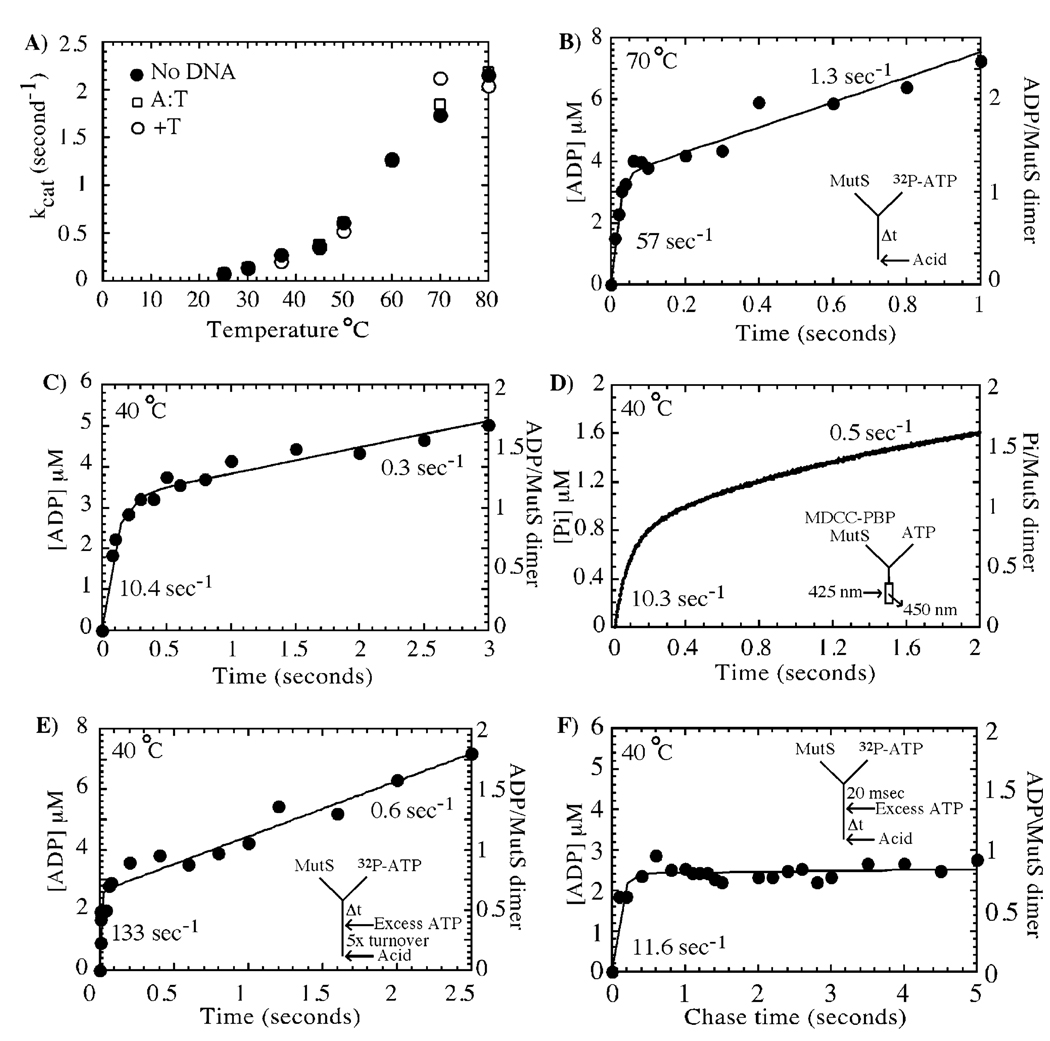
The asymmetry in ATP or ADP binding to the MutS dimer raises the possibility that the two subunits might also hydrolyze ATP in an asymmetric manner. To test this hypothesis, we decided to directly measure the ATP binding, hydrolysis, and product release steps in one catalytic turnover of the MutS ATPase. Initially, steady-state experiments were performed to determine the optimal reaction conditions for such measurements. As reported previously, T. aquaticus MutS ATPase activity increases with temperature (42), and as shown in Figure 2A, the kcat approaches a high value of 2 s−1 at 80 °C. The steady-state ATPase activity also exhibits salt dependence and is maximal at 150 mM KCl (data not shown). Thus, initial rapid quench experiments measuring ATP hydrolysis under pre-steady-state conditions were performed at 70 °C and with 150 mM KCl in the reaction buffer. The data in Figure 2B show a MutS-catalyzed burst of ADP formation at 57 s−1, followed by a linear phase at 1.3 s−1 (velocity/[MutSdimer]), in the absence of any DNA substrate. Thus, MutS hydrolyzes ATP rapidly, but catalytic turnover is limited by a slow step in the reaction following ATP hydrolysis. Interestingly, the amplitude of the burst phase is 3.5 µM, which is consistent with rapid ATP hydrolysis occurring at only one subunit of the MutS dimer (3 µM dimer = 6 µM monomers or ATPase sites).
FIGURE 2.
Asymmetry in ATP binding is recapitulated during ATP hydrolysis. In panel A, steady-state ATPase activity of T. aquaticus MutS is temperature-dependent with kcat approaching a maximum of 2 s−1 at 80 °C. In panel B, pre-steady-state rapid-quench analysis of MutS activity at 70 °C reveals a burst of ATP hydrolysis at a rate of 57 ± 11 s−1 and amplitude of 3.5 ±0.15 µM, followed by a slow steady-state phase at 1.3 ± 0.12 s−1 (3 µM MutS dimer and 500 µM ATP in the reaction). The burst phase is equivalent to rapid hydrolysis of one ATP molecule per MutS dimer. In panel C, the burst rate and kcat vary with temperature (e.g., at 40 °C the burst rate constant is 10.4 s−1 and the linear rate constant is 0.3 s−1, although the amplitude is still 3.2 µM, see Table 1). In panel D, a fluorescent reporter assay measures a burst of phosphate release from 1 µM MutS at 10.3 ± 0.3 s−1 at 40 °C and 1.1 ± 0.02 µM amplitude (one ADP/Pi per MutS dimer), suggesting that the phosphate is released rapidly following ATP hydrolysis. In panel E, pulse–chase experiments at 40 °C with 3 µM MutS and 500 µM ATP also show a burst amplitude of one ADP per dimer and yield an ATP binding rate constant of 0.25 ± 0.1 µM−1 s−1. In panel F, a pulse–chase time experiment, in which hydrolysis of 32P-ATP (500 µM) bound to MutS (3 µM dimer) prior to addition of unlabeled ATP chase (10 mM) is observed over time, detects only one ADP formed per MutS dimer in the first turnover (burst rate = 11.6 ± 2.3 s−1).
The ATPase assays were repeated under various conditions to determine any effects of salt and temperature on the burst amplitude and rate of ATP hydrolysis. Under all KCl concentrations tested (0–150 mM), rapid hydrolysis of only one ATP per MutS dimer was observed, followed by a 30–50-fold slower steady-state ATPase rate (data not shown). Consistent with the steady-state data, both the burst rate and the slow turnover rate increased with increasing temperature; however, the amplitude of the burst phase remained constant at one ADP formed per MutS dimer at all temperatures tested above 25 °C (Table 1). To measure reaction rates accurately (at rates slower than ~60 s−1) and to maintain the integrity of the quench-flow apparatus, further pre-steady-state experiments were carried out at 40 °C (Figure 2C, burst rate constant = 10.4 s−1, burst amplitude = 3.2 µM, steady-state kcat = 0.3 s−1).
Table 1.
Temperature Dependence of MutS Pre-Steady-State Parameters
| temperature (°C) | burst amplitude (µM) (3 µM = one site) |
burst rate (s−1) | kcat (s−1) |
|---|---|---|---|
| 25 | 2.1 ± 0.17 | 2.9 ± 0.49 | 0.3 ± 0.05 |
| 40 | 3.2 ± 0.07 | 10 ± 1.15 | 0.3 ± 0.01 |
| 50 | 3.5 ± 0.11 | 23 ± 4.8 | 0.6 ± 0.08 |
| 60 | 2.9 ± 0.21 | 33 ± 8.2 | 0.7 ± 0.15 |
| 70 | 3.5 ± 0.15 | 57 ± 11.2 | 1.3 ± 0.12 |
The burst kinetics of ATP hydrolysis observed in rapid quench experiments indicate that a posthydrolysis step (or steps), perhaps related to product release, is responsible for limiting the catalytic turnover rate. We measured the rate of phosphate (Pi) release using a real-time Pi-release assay developed by the Webb research group (38). MDCC-labeled phosphate binding protein (MDCC–PBP) was used as a fluorescent reporter of Pi in solution; PBP binds Pi rapidly (1.4 × 108 M−1 s−1) and with high affinity (Kd = 0.1 µM), and its fluorescence increases ~7-fold upon interaction with Pi (data not shown; ref 43). Figure 2D shows a stopped-flow trace of the increase in MDCC–PBP fluorescence over time as MutS (1 µM dimer) hydrolyzes ATP and releases the Pi product at 40 °C. Calibration curves relating the fluorescence signal to known Pi concentrations were used to quantitate the amount of Pi released during the time course (data not shown). The data reveal an exponential burst of one Pi released per MutS dimer at 10 s−1, followed by a slow linear phase at 0.5 s−1. The Pi release rate appears identical to the ATP hydrolysis rate measured in Figure 2C, indicating that after ATP hydrolysis Pi dissociates very rapidly from MutS and that a step after Pi release—perhaps associated with ADP dissociation or perhaps hydrolysis of ATP at the second subunit—is responsible for the slow turnover.
Next, pulse–chase experiments were carried out to characterize the kinetics of ATP binding to MutS. The protein was mixed with α32P-ATP for varying times and chased with 20-fold excess unlabeled ATP for time equivalent to 5–6 turnovers. During the chase, bound α32P-ATP may be hydrolyzed to α32P-ADP and Pi or remain unhydrolyzed—either bound to MutS or released into solution. Any free α32P-ATP in solution is diluted upon addition of unlabeled ATP chase and is not available for binding and hydrolysis. Thus, the pulse–chase assay measures the rate of ATP binding to MutS and the fraction of the MutS•ATP complex that undergoes hydrolysis. The data in Figure 2E reveal a burst of ADP formation at 133 s−1 with 2.8 µM amplitude (3 µM MutS dimer and 500 µM ATP in the reaction). The burst rate increases linearly with ATP concentration and yields an apparent bimolecular rate constant of 2.5 × 105 M−1 s−1 for ATP binding to MutS. The burst amplitude remains constant at one ADP formed per MutS dimer, affirming results from acid quench experiments indicating that only one subunit of the MutS dimer catalyzes rapid ATP hydrolysis. The second subunit may (a) not bind ATP, (b) hydrolyze ATP at a slow rate that is indistinguishable from the steady-state rate, or (c) retain or release ATP unhydrolyzed. According to the Kd values measured in Figure 1, 500 µM ATP in the reaction is likely sufficient to saturate binding to both subunits of the MutS dimer; thus, we favor the possibility that a second ATP is bound but hydrolyzed slowly or not hydrolyzed within one turnover (the possibility of a high rate of reverse chemistry, that is, ATP resynthesis, is under investigation).
Given the above indications of asymmetry in ATPase activity of the MutS dimer, we were intrigued by the possibility that the two subunits might hydrolyze ATP sequentially; that is, ATP hydrolysis at the second subunit follows hydrolysis at the first subunit. To address this question, the pulse–chase experiment described above was modified to observe any slow ATP hydrolysis that might occur within the first turnover. In this “chase time” experiment, 3 µM MutS dimer was first mixed with 500 µM α32P-ATP for 20 ms to allow α32P-ATP binding to MutS (binding rate constant = 2.5 × 105 M−1 s−1, Figure 2E), then mixed with 10 mM unlabeled Mg2+ ATP, and the reaction was allowed to continue for varying chase times, followed by acid quench. During the chase time, we can observe formation of α32P-ADP from all α32P-ATP bound to MutS during the binding period that undergoes hydrolysis prior to dissociation (no α32P-ATP can bind MutS during the chase time due to dilution with excess unlabeled ATP). Figure 2F shows a single-exponential burst of ADP formation at 11 s−1, as expected from the results of the rapid quench experiment in Figure 2C. The burst phase saturates at 2.5 µM ADP or one ADP per MutS dimer, and there is no evidence of another exponential phase as expected if the second subunit hydrolyzed α32P-ATP at 0.3–0.4 s−1 following the first one (similar results were obtained even when the initial binding period was extended to 200 ms to provide more time for the second ATP to bind MutS; data not shown). These data reveal that the second MutS subunit does not hydrolyze ATP within the same catalytic turnover as the first subunit. ATP bound to this site may be retained unhydrolyzed (noncatalytic site), or it may be bound weakly such that during the chase time α32P-ATP dissociates from MutS faster than it can be hydrolyzed to α32P-ADP. In the latter case, the second subunit would be catalytically active, but the reaction would appear slow due to a weak ATP binding equilibrium. In either case, though, the ATPase activity of the two MutS subunits does not appear to be sequentially linked in that ATP hydrolysis at one subunit does not trigger hydrolysis at the other.
Rapid ATP Hydrolysis Occurs at the High-Affinity Nucleotide-Binding Site on the MutS Dimer
We investigated MutS ATP binding and hydrolysis activities further to determine whether the second subunit in the dimer is catalytically active. The first step in this process was to identify which of the two MutS subunits catalyzes rapid ATP hydrolysis. The identical data obtained from rapid-quench assays (Figure 2C) and MDCC–PBP-based Pi reporter assays (Figure 2D) demonstrated that Pi release is an accurate and relatively simple measure of MutS ATP hydrolysis activity. Pi release kinetics measured at increasing concentrations of ATP (5–1000 µM, Figure 3A) show the burst rate constant increasing in a hyperbolic manner to a maximum of 10.7 s−1 with a K1/2 of 33 µM (Figure 3B). This K1/2 value is very close to the Kd of 27 and 33 µM measured for ATPγS and ADP binding, respectively, to the low-affinity nucleotide-binding site on MutS (Figure 1). A straightforward interpretation of this coincidence is that the low-affinity site catalyzes the rapid burst of ATP hydrolysis. However, this K1/2 can only be considered as a kinetic measure of Kd (akin to a Michaelis constant), which may not necessarily reflect the true dissociation constant for the MutS–ATP interaction (e.g., if the binding is not in rapid equilibrium). More importantly, a plot of the burst amplitude versus ATP concentration yields a K1/2 = 2.8 µM (Figure 3C). Because of greater than 30-fold difference between the rates of product formation and turnover (Figure 2B,C), the ATP dependence of burst amplitude reflects an active site titration, and the 2.8 µM K1/2 is a measure of the Kd for the active site. This value is consistent with the low Kd values measured for ATP, ATPγS, and ADP binding to the high-affinity site on MutS (Figure 1) and therefore favors a model in which the high-affinity site catalyzes rapid ATP hydrolysis. As noted above, the high K1/2 value in Figure 3B could reflect ATP binding as well as other steps in the reaction, although it could also indicate that ATP binding to the low-affinity site (with Kd ≈ 30 µM) is coupled to ATP hydrolysis at the high-affinity site (at this time there is no evidence to support or discount the latter hypothesis).
To confirm that the high-affinity site catalyzes rapid ATP hydrolysis, we measured the reaction (Pi release) with MutS preincubated with low concentrations of ATPγS (1–10 µM), which should occupy the high-affinity nucleotide-binding site and block its activity (Kd = 3 µM for ATPγS binding to the high-affinity site, Figure 1A). A representative trace at 10 µM ATPγS concentration shows that the burst of ATP hydrolysis is suppressed completely when the activity of the high-affinity site is blocked by ATPγS (Figure 3D). A plot of the decrease in burst amplitude versus ATPγS concentration fit to a hyperbola yields K1/2 = 2.5 µM (Figure 3E), which is in good agreement with the Kd of 3 µM measured for ATPγS binding to the high-affinity nucleotide-binding site on the MutS dimer (Figure 1A). Thus, these data also support the hypothesis that the ATP molecule bound tightly to MutS undergoes rapid hydrolysis in the absence of DNA (in the following text, we designate the high-affinity, rapid hydrolysis site as “S1” and the other site as “S2”).
Slow ATP Hydrolysis Occurs at the Low-Affinity Nucleotide-Binding Site on the MutS Dimer
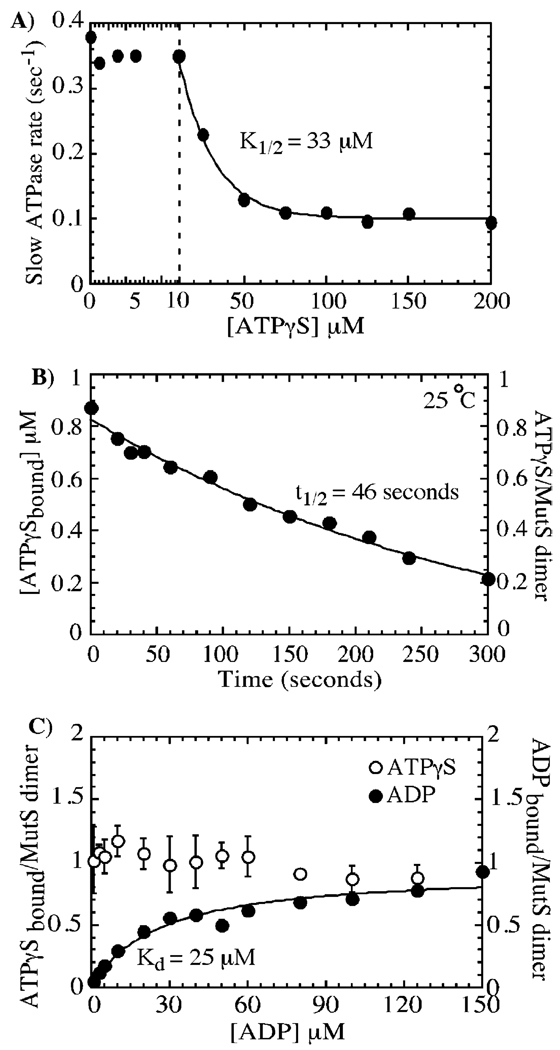
Another interesting finding from the experiments described above is that while low ATPγS concentrations suppress the burst phase of ATP hydrolysis, the linear ATPase phase remains unchanged at a 0.3 s−1 rate (Figure 3D, trace with 10 µM ATPγS shown). This raises the possibility that the low-affinity nucleotide-binding site (S2) is catalytically active and continues to hydrolyze ATP even as activity of the high-affinity site (S1) is blocked by ATPγS. Further increases in ATPγS concentration do suppress the slow ATPase rate in a hyperbolic manner, and a plot of the decreasing rate constants versus ATPγS concentration yields a K1/2 of 33 µM (Figure 4A). This value is again very similar to the Kd of 27 µM measured for ATPγS binding to S2 (Figure 1A) and indicates that the slow ATPase rate reflects catalytic activity of the low-affinity nucleotide-binding site on the MutS dimer.
FIGURE 4.
The low-affinity site on ATPγS-bound MutS hydrolyzes ATP slowly, yielding an ATPγS–ADP-bound MutS dimer species. In panel A, further analysis of the data in Figure 3 shows that inhibition of the slow ATPase phase (rate constant = 0.3–0.4 s−1) requires higher ATPγS concentrations, and the plot of rate constants versus ATPγS fit to a hyperbola yields K1/2 = 33 µM (similar to the Kd of 27 µM for ATPγS binding to the low-affinity site on MutS, see Figure 1A); ATPase data for 150 µM ATPγS are shown in Figure 3D. In panel B, a nitrocellulose filter-binding experiment, in which 35S-ATPγS (12 µM) bound to MutS (1 µM dimer) is chased by ATP (500 µM), shows that ATPγS dissociates from MutS at a very slow rate of 0.015 s−1 (t1/2 = 46 s); that is, it remains bound to the MutS high-affinity site during the slow ATPase phase. In panel C, MutS (1 µM dimer) preincubated with 35S-ATPγS (○; 5 µM) and titrated with 32P-ADP (●; 0–150 µM) shows binding of one ADP molecule to the low-affinity site (Bmax = 0.93 ± 0.06 µM and Kd = 25 ± 5 µM) and retention of the ATPγS molecule already bound to the high-affinity site.
Figure 4B shows the rate of dissociation of a tightly bound 35S-ATPγS (12 µM in the reaction) from MutS when chased with excess unlabeled ATP (500 µM), measured by nitrocellulose membrane filtration. The long half-life of the complex (46 s, koff = 0.015 s−1) confirms that in the ATP hydrolysis (Pi release) experiments described here ATPγS remains bound to the S1 site even as the S2 site continues to hydrolyze ATP (2-s time course, Figure 3D and Figure 4A) (note that the ATPγS dissociation rate was measured at 25 °C due to limitations with using nitrocellulose membranes at 40 °C, whereas the ATPase data shown here were measured at 40 °C; however, identical ATPase assays performed at 25 °C also show ongoing ATP hydrolysis at 0.2 s−1 after the S1 site is occupied by nonhydrolyzable ATPγS, data not shown).
Continued ATP hydrolysis at one subunit of the MutS dimer while the other subunit remains ATP-bound predicts a MutS form that can bind both ATP and ADP simultaneously. The experiment in Figure 4C tests this hypothesis by titrating 1 µM MutS dimer containing a tightly bound ATPγS molecule with increasing concentrations of ADP. The 35S signal shows that the ATPγS remains bound to MutS even with 150 µM ADP added to the reaction, and the 32P signal shows that one ADP molecule binds MutS•ATPγS complex with a Kd of 25 µM, as expected for nucleotide binding to the low-affinity S2 site (Figure 1B).
A +1 Insertion-Containing DNA Suppresses MutS ATPase Activity
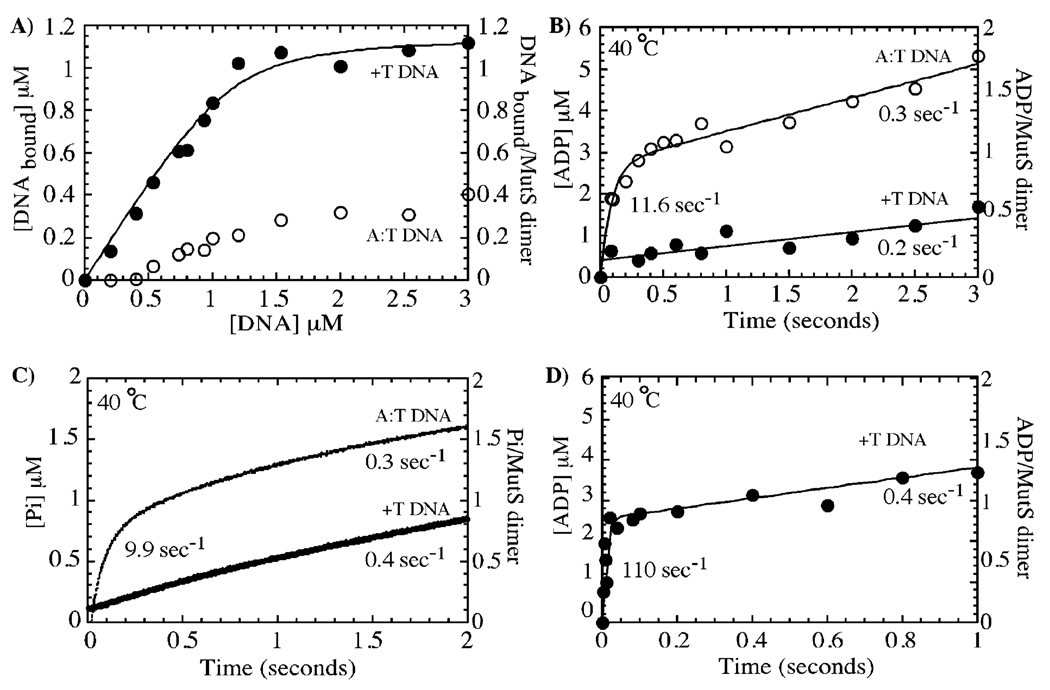
The data in Figure 3 and Figure 4 identified asymmetric rapid and slow ATP-hydrolyzing sites in the MutS dimer and indicated possible occurrence of an S1ATP–S2ADP species during the ATPase reaction. The next series of experiments examine the relevance of these findings for MutS DNA binding and mismatch recognition activities. Membrane filter assays show that MutS binds a +T insertion-containing DNA duplex with 1:1 stoichiometry and with higher affinity and stability than fully matched DNA, consistent with previous reports of selective MutS interaction with DNA mismatches or insertions (Figure 5A, sequence copied from the T. aquaticus MutS–DNA crystal structure, ref 14) (1, 42, 44–46). Figure 5B shows MutS ATPase activity in the presence of the two DNA substrates. In the presence of the A:T matched duplex, the MutS ATPase profile is indistinguishable from that in the absence of DNA with a burst phase at a rate constant of 11.6 s−1 and amplitude of 2.7 µM (3 µM MutS dimer) followed by a linear phase at 0.3 s−1. These data suggest that MutS binding to fully matched DNA does not affect its ATPase activity, but given that the interaction is weak, it is also possible that MutS exists predominantly as free protein in the presence of fully matched DNA. In stark contrast to the above results, the presence of +T DNA in the reaction leads to complete suppression of the burst of ATP hydrolysis, and only the linear phase is detectable at 0.2 s−1 (Figure 5B). Phosphate release experiments confirm that MutS ATPase activity is unchanged in the presence of A:T DNA but is suppressed by +T DNA and show that Pi release remains faster than ATP hydrolysis even after MutS binds DNA (Figure 5C, apparent Pi release rate is the same as the ATP hydrolysis rate). Thus, +T DNA binding to MutS changes its ATPase mechanism such that a step before or at ATP hydrolysis becomes rate-limiting. The MutS•+T DNA complex may bind ATP more slowly or weakly, resulting in slow ATP hydrolysis, or it may bind ATP rapidly (as in the absence of DNA) and then undergo a delay in hydrolysis. A pulse–chase experiment carried out in the presence of +T DNA shows that at least one ATP molecule binds rapidly to the MutS•+T DNA complex (Figure 5D, binding rate constant = 2.2 × 105 M−1 s−1), just as in the absence of DNA (Figure 2E, binding rate constant = 2.7 × 105 M−1 s−1), and is hydrolyzed during the time of chase. Thus, the slow step in the MutS•+T DNA ATPase pathway occurs after ATP binding and before or at ATP hydrolysis.
FIGURE 5.
Mismatched DNA suppresses ATP hydrolysis. In panel A, nitrocellulose membrane filtration experiments with 1 µM MutS dimer titrated with 32P-DNA show near 1:1 stoichiometric interaction between MutS and DNA containing a +T insertion but significantly weaker interaction with fully matched DNA. In panel B, preincubation of MutS (3 µM dimer) with mismatched DNA (5 µM) results in complete inhibition of the burst of ATP hydrolysis (●); ATP hydrolysis in the presence of fully matched DNA (○; burst rate constant = 11.6 ± 2 s−1 and amplitude = 2.7 ± 0.1 µM or one ADP per MutS dimer) is the same as in the absence of DNA (see Figure 2C). In panel C, phosphate-release kinetics of MutS (1 µM dimer) confirm that ATP hydrolysis is suppressed by +T DNA and not by fully matched DNA and that the phosphate-release rates remain fast even in the presence of DNA. In panel D, pulse–chase analysis reveals rapid ATP binding to the MutS•+T DNA complex at 0.22 µM−1 s−1 followed by slow hydrolysis, confirming that mismatched DNA specifically inhibits the ATP hydrolysis step.
A +1 Insertion-Containing DNA Suppresses ATP Hydrolysis Specifically at the High-Affinity Nucleotide-Binding Site on MutS
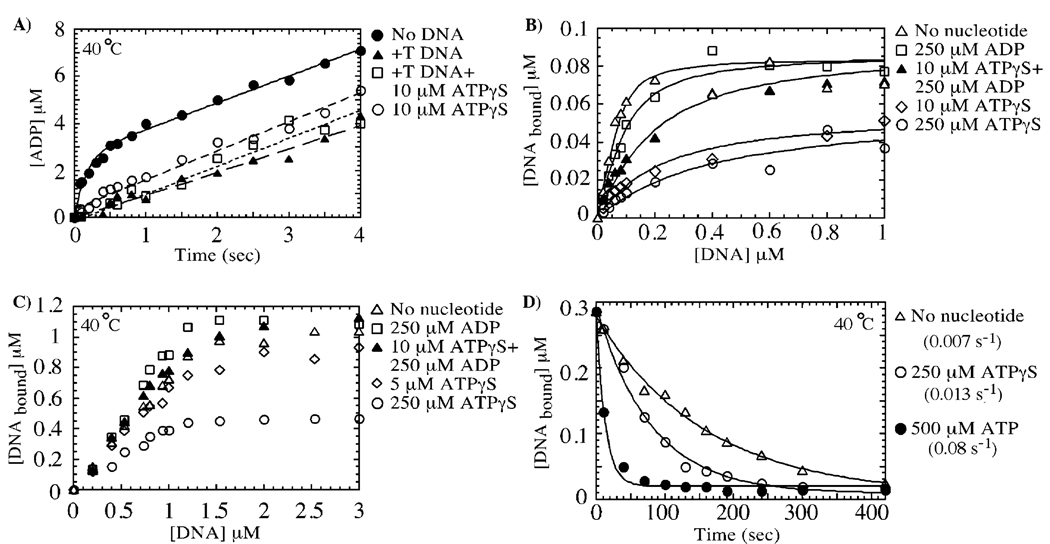
In addition to revealing the inhibitory effect of +T DNA on MutS-catalyzed rapid ATP hydrolysis, the data in Figure 5B show that the linear ATPase phase continues at a rate of 0.2 s−1. Thus, the question arises whether this ATPase activity is from the +T-inhibited S1 site (implying that it now hydrolyzes ATP at a much slower rate or after slow dissociation of +T DNA from MutS) or from the S2 site (implying that +T DNA does not inhibit the S2 ATPase). To address this issue, ATPase experiments were performed with MutS in the presence of both +T DNA and ATPγS. As shown previously in Figure 3, MutS alone exhibits a burst of rapid ATP hydrolysis followed by a slow linear phase (Figure 6A). +T DNA and 10 µM ATPγS each have very similar effects on MutS ATPase activity, that is, suppression of the burst phase but no obvious effect on the linear phase (Figure 3D, Figure 5B, and Figure 6A). Interestingly, MutS in the presence of both 10 µM ATPγS and +T DNA also shows the same kinetic profile (Figure 6A; linear rate constant = 0.33 s−1). Under these conditions, ATPγS is bound stably to the high-affinity S1 site and its activity is blocked; two ATPγS molecules bind MutS•+T DNA with Kd = 10 and 75 µM, and half-life of ATPγS on the S1 site of MutS is 50 s, just as in the absence of DNA (Figure 1A and Figure 4B, data with DNA not shown). Thus, continuing ATP hydrolysis at 0.3 s−1 indicates that +T DNA does not inhibit the slow ATPase activity of the low-affinity S2 site.
FIGURE 6.
Mismatched DNA inhibits ATP hydrolysis specifically at the high-affinity site, yielding an ATP/ADP-bound form of MutS, which has high affinity for mismatched DNA. In panel A, +T DNA binding to MutS (3 µM dimer) suppresses the burst of ATP hydrolysis (▲), but the slow phase remains identical to that in the absence of DNA (●, 0.33 s−1). The same slow ATPase rate is observed for MutS preincubated with 10 µM ATPγS (○, high-affinity site occupied, no burst phase) or with 10 µM ATPγS and +T DNA (□), indicating that the low-affinity site continues to hydrolyze ATP when MutS is bound to mismatched DNA. Panels B and C show nitrocellulose membrane filtration experiments of 0.1 and 1 µM MutS dimer, respectively, binding to +T DNA in the absence of nucleotide cofactors (∆, Kd = 20 nM), in the presence of 250 µM ADP (□, both high-affinity and low-affinity sites occupied, Kd = 45 nM), and in the presence of 10 µM ATPγS and 250 µM ADP (▲, ATPγS in high-affinity site and ADP in low-affinity site, Kd = 100 nM). The interaction between MutS and +T DNA is weaker in the presence of 10 µM ATPγS (◊, one site occupied by ATPγS, Kd = 200 nM; note stable +T DNA interaction with this species at 1 µM MutS) and 250 µM ATPγS (○, both sites occupied with ATPγS, Kd ≈ 300 nM; note weak +T DNA interaction with this species even at 1 µM MutS). In panel D, the rate of dissociation of 32P-labeled +T DNA (1 µM) from MutS (0.5 µM) was measured over time by membrane filtration assays using 50 µM unlabeled +T DNA chase (with or without nucleotides). The dissociation rate is 0.007 s−1 in the absence of nucleotides (∆), 0.013 s−1 in the presence of 250 µM ATPγS (○), and 0.077 s−1 in the presence of 500 µM ATP (●).
+T DNA stabilizes S1 in an ATP-bound state, while slow ATP hydrolysis continues at S2, implying that the MutS•+T DNA complex can exist in relatively long-lived S1ATP–S2ATP and S1ATP–S2ADP forms in the ATPase pathway. We then questioned the relevance of these nucleotide-bound MutS forms in the DNA mismatch repair pathway. In Figure 6B, data from membrane filtration experiments in with 0.1 µM MutS show that S10–S20 (Kd = 20 nM) and S1ADP–S2ADP (Kd = 45 nM) forms bind +T DNA tightly, and S1ATPγS–S2ADP binds the mismatch with lower affinity than the above species (Kd = 100 nM) but more tightly than do S1ATPγS–S20 (Kd ≈ 200 nM) and S1ATPγS–S2ATPγS (Kd ≈ 300 nM); note that only about 50% DNA binding is detectable with these latter two MutS species. At a higher protein concentration (1 µM MutS dimer), most of the nucleotide-bound MutS species, including S1ATPγS–S2ADP and even S1ATPγS–S20, form stable, near 1:1 stoichiometric complexes with +T DNA (Figure 6C). Only S1ATPγS–S2ATPγS continues to exhibit weak, unstable interaction with +T DNA, similar to that between MutS and fully matched DNA (compare Figure 5A and Figure 6C).
According to the above results, many intermediate species in the MutS ATPase pathway can bind +T DNA with differing but relatively high affinities except for MutS with both subunits occupied by ATP. However, even this form of MutS may maintain a fairly stable interaction with DNA as indicated by the 50-s-long half-life of MutS•+T DNA complex in the presence of 250 µM ATPγS (koff = 0.013 s−1, Figure 6D) or the 10-s half-life of MutS•+T DNA in the presence of 500 µM ATP (koff = 0.077 s−1; Figure 6D). Rates of DNA dissociation from these MutS species were measured at 40 °C by preincubating MutS with 32P-DNA, followed by simultaneous addition of 50-fold excess unlabeled +T DNA chase and the relevant nucleotide (e.g., 250 µM ATPγS yields the S1ATPγS–S2ATPγS species) and membrane filtration at varying times (Figure 6D). S10–S20•+T DNA is clearly the most stable complex with a half-life of 100 s (koff = 0.007 s−1). The stability of this complex is lowered substantially (10-fold) upon ATP binding and formation of S1ATP–S2ATP; nevertheless, the 0.077 s−1 dissociation rate of +T from S1ATP–S2ATP is slower than or comparable to the rate of ATP hydrolysis at S2 (0.3 s−1, Figure 6A) and formation of the S1ATP–S2ADP species that binds +T DNA with fairly high affinity (100 nM, Figure 6B). We therefore consider it quite possible that during the ATPase reaction, even as MutS binds two ATP molecules and loosens its hold on mismatched DNA, it hydrolyzes ATP at the low-affinity nucleotide-binding site and becomes capable of rebinding the DNA.
DISCUSSION
In recent years, the efforts of several research groups have yielded overwhelming evidence for nonequivalence of DNA binding and ATPase activities of the two subunits in the dimeric MutS DNA mismatch repair protein (18, 29–33). For example, crystal structures of both T. aquaticus and E. coli MutS proteins show that a phenylalanine from only one subunit stacks against an unpaired or mispaired base in the duplex (14, 15), and the E. coli MutS structure shows an ADP molecule bound to only one subunit (15). Consistent with this structural data, the Modrich research group detected only one ADP, AMPPNP, or ATPγS molecule binding per E. coli MutS, as well as human Msh2–Msh6, dimer (29, 47). At the same time, our study of S. cerevisiae Msh2–Msh6 detected only one ATP or one ADP binding per heterodimer, and we also demonstrated that two ATPγS molecules bind the dimer with differing affinities (3–5-fold difference, ref 31). Moreover, all these MutS proteins are capable of binding one ATPγS and one ADP molecule simultaneously. Another key finding of our study was that Msh2–Msh6 appears to hydrolyze only one ATP molecule in a catalytic turnover, even though both Msh2 and Msh6 proteins possess the Walker A and B motifs for ATPase activity. These results further solidified speculation that the two subunits in a MutS dimer do not bind or hydrolyze ATP in an identical fashion and predicted multiple nucleotide-bound species in the MutS ATPase pathway (29, 31). Identification these nucleotide-bound species and their roles in the ATPase-coupled DNA mismatch repair activity of MutS has become an exciting avenue of current research.
Having chosen to focus on MutS ATPase activity initially, we took the approach of quantitatively measuring ATP binding, hydrolysis, and product release events in real time to explicitly determine the origin and consequences of asymmetry in the two MutS subunits during mismatch repair. First, corroborating and extending the studies of S. cerevisiae Msh2–Msh6 and E. coli MutS proteins detailed above, we observed differential binding of two ATPγS molecules and even two ADP molecules per T. aquaticus MutS dimer (note that only one ATP appears to be bound per dimer, but it is likely that the second binding is too weak for detection in the membrane filtration assay). An important caveat to consider when such differential activity is observed is that the protein—a recombinant protein produced in E. coli, at that—may exist in multiple populations with varying conformations and activities. It is difficult to conclusively rule out this possibility; however, our concerns were eased by the “half-site” nucleotide binding observed for E. coli MutS and human Msh2–Msh6 (29, 47) and by very similar results with S. cerevisiae Msh2–Msh6 as well (31). Given that proteins from four different species, analyzed in two different laboratories, exhibit tight binding of one nucleotide and weak (sometimes undetectable) binding of a second nucleotide per MutS dimer, it seems quite unlikely that these results are a consequence of mixed protein populations.
Upon measuring T. aquaticus MutS ATPase activity under pre-steady-state conditions, we again observed “half-site” reactivity, in that only one ATP molecule was bound and hydrolyzed rapidly prior to a slow, rate-limiting step in the ATPase reaction. As argued earlier, the possibility that the asymmetry in T. aquaticus MutS ATPase activity reflects a mixture of protein populations is low, given identical results with the S. cerevisiae Msh2–Msh6 heterodimer (31).
Asymmetric Equals Sequential?
If only one subunit in the dimer hydrolyzes ATP rapidly, what happens at the other subunit? Is ATP bound to it? If so, is the ATP hydrolyzed? If so, when is it hydrolyzed? Are the two subunits coupled to hydrolyze ATP sequentially? Answers to these questions are necessary to identify nucleotide-bound forms of MutS that occur in the ATPase and mismatch repair pathway. The 10-fold difference in nucleotide-binding affinity between the two subunits of T. aquaticus MutS provided us with an opportunity to observe the activity of the low-affinity subunit (S2) while the activity of the high-affinity subunit (S1) was blocked by ATPγS (in case of S. cerevisiae Msh2–Msh6, the difference was only ~3-fold, making it difficult to block one subunit selectively, ref 31). Consequently, we were able to determine that S1 hydrolyzes ATP rapidly, at 10 s−1, and S2 appears to hydrolyze ATP slowly, at 0.3 s−1 (40 °C).
To address the question of sequential ATPase activity, we attempted to follow ATP hydrolysis at the second subunit by the “chase time” experiment described in Figure 2F. First, MutS protein was given sufficient time to bind radiolabeled ATP, and then 20-fold excess unlabeled ATP was added to the reaction to prevent further labeled nucleotide binding to the protein. The chase was continued for a time, and the reaction was sampled at regular intervals to detect hydrolysis of labeled ATP molecules bound to MutS during the initial binding period. As expected, we observed rapid formation of one ADP molecule, but after that no further reaction could be detected. Since we have evidence that S2 is catalytically active but it binds nucleotides with low affinity, we conclude that labeled ATP bound to S2 dissociates during the chase time, prior to hydrolysis. S2 likely hydrolyzes ATP over the course of multiple ATP binding/release events, and the ATPase rate might appear slow due to the unfavorable ATP binding equilibrium. These results indicate that the catalytic activity of S2 is not sequentially linked to that of S1. In fact, S2 appears capable of hydrolyzing ATP even when the S1 subunit is rendered inactive (Figure 3D and Figure 4A).
Steady-state ATPase analyses have been employed previously to determine whether and how ATPase activities of the two MutS subunits are coupled to each other. In one study, incubation of E. coli MutS with vanadate (Vi) was found to suppress the steady-state ATPase rate from 12.5 µM ADP/min to 2.7 µM ADP/min (30). Viewed in conjunction with data suggesting that one MutS subunit can be trapped in an ADP–Vi state, the Vi-induced drop in ATPase activity was interpreted as inhibition of ATP hydrolysis at the second MutS subunit after the first subunit was trapped in an ADP–Vi state. Additionally, the report included a version of the “chase time” experiment described here, in which the authors initiated an unlabeled ATP chase during steady state and observed additional hydrolysis of labeled ATP during the time of chase (in contrast, we detect no further activity with Taq MutS following one ATP hydrolysis, Figure 2F). These results led the authors to conclude that the MutS subunits catalyze ATP hydrolysis in an alternating or sequential fashion. However, the pre-steady-state analyses of MutS ATPase activity shown here—and previously for S. cerevisiae Msh2–Msh6 (31) and E. coli MutS (34)—have revealed that the steady-state ATPase rate does not reflect the rate of ATP hydrolysis, but rather a step or steps following the hydrolysis event. In this context, it is also possible to interpret the Vi-inhibition data as follows: the second subunit could catalyze ATP hydrolysis at any rate that is faster, equal to, or slower than the rate-limiting step in the ATPase pathway of the first subunit and it would not be distinguishable from the steady-state rate; when the first subunit is blocked in an ADP–Vi state, the ATPase rate at the second subunit could become measurable, but it is not possible to determine from this experiment whether the measured rate represents normal, inhibited, or even stimulated ATPase activity at the second subunit. Regarding the results from the “chase time” assay, the difference in amplitude with and without chase is very small (due to low concentration of protein used in the experiment), also only two data points were collected following addition of chase, and finally, the use of EDTA as a quenching agent may lead to anomalous results, since EDTA only sequesters free Mg2+ and may in fact serve as a chase rather than an instantaneous quench of the ATPase reaction (Hingorani, M. M., and Patel, S. S., unpublished results, ref 39). Given these caveats and possible alternative interpretations of the steady-state ATPase data, it is premature to invoke a coupled sequential ATPase mechanism for the MutS subunits at this time, because it may lead to incorrect identification of MutS species in the ATPase pathway and corresponding incorrect hypotheses regarding their function in DNA mismatch repair.
Thus far we have conclusive evidence for asymmetric and possibly partially coupled ATP binding and hydrolysis activities of the two MutS subunits (ATP binding to S2 may be associated with rapid ATP hydrolysis at S1, Figure 3B). It remains possible that the S1 subunit cannot turn over after ATP hydrolysis until S2 hydrolyzes ATP as well, but thus far there is no direct evidence to support this type of sequential ATPase mechanism.
Several Nucleotide-Bound Forms of MutS Occur in the DNA Mismatch Repair Pathway
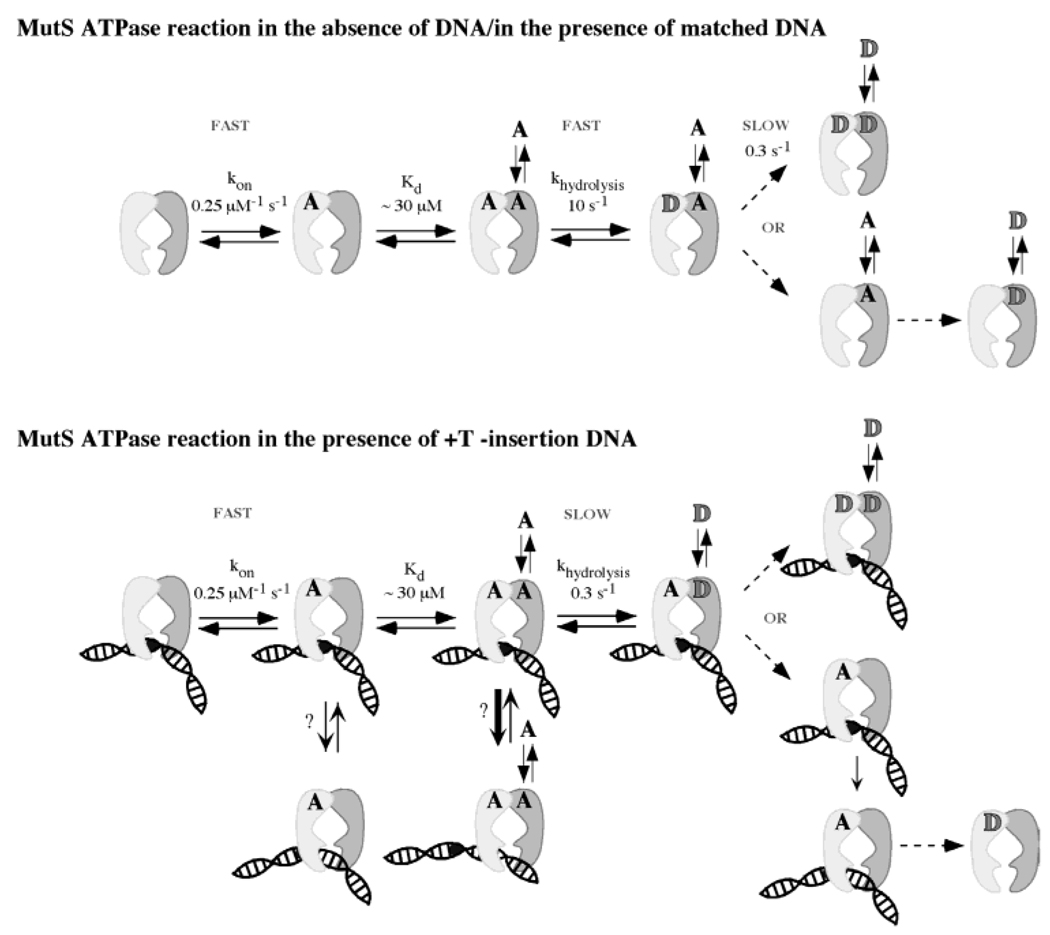
The ATPase reaction catalyzed by MutS bound to mismatched DNA is very different from that of free MutS with the catalytic activity of S1 subunit suppressed while S2 remains active. The results from our kinetic measurements have been summarized in Figure 7 in model pathways that also depict various forms of MutS that occur during the reaction in the absence or presence of mismatched DNA.
FIGURE 7.
Prominent species in the MutS ATPase pathway and their interactions with DNA. According to the kinetic parameters measured for MutS ATPase, in the absence of DNA or in the presence of fully matched DNA, the first slow, rate-limiting step occurs after ATP hydrolysis at S1, indicating that S1ADP–S2ATP might be a stable species in this pathway. In contrast, when MutS is bound to a DNA mismatch, the first rate-limiting step occurs prior to ATP hydrolysis at S1, indicating that S1ATP–S2ATP and perhaps S1ATP–S2ADP as well might be the stable species in this ATPase pathway. The model also depicts possible interactions between various nucleotide-bound MutS species and mismatched DNA based on results from DNA-binding experiments in this study and in the literature. Dashed arrows indicate steps that may not happen in immediate succession.
In the absence of DNA, the first slow step in the ATPase reaction occurs following ATP hydrolysis at S1, which suggests that the S1ADP–S2ATP (or S1ADP–S20) species has a prolonged lifetime in the pathway. When MutS is bound to DNA containing a single +T insertion, the first slow step in the reaction occurs prior to ATP hydrolysis at S1, which suggests that the S1ATP–S2ATP species—or S1ATP–S2ADP and S1ATP–S20 species or both, given continuing slow ATP hydrolysis and weak ATP or ADP binding at S2—have a prolonged lifetime in the pathway. The predominance of distinctly different species in the ATPase reaction following MutS binding to mispaired/unpaired bases as compared with free MutS implies that these species perform key functions in DNA mismatch repair.
What Is Their Function?
As noted earlier, nucleotide binding to MutS proteins induces changes in their conformation and modulates their interactions with DNA. Several research groups have reported an apparent reduction in the affinity of MutS for mismatched DNA following ATP binding, which manifests as MutS sliding off short linear mismatched DNA substrates with unblocked ends during gel mobility-shift analysis (1, 13, 23–25, 28, 48); presumably, such movement of MutS on DNA serves an important function during mismatch repair. Since we have now identified some nucleotide-bound forms of MutS in the pathway, we tested their interaction with +T-containing duplex DNA substrate. Consistent with previous studies (21), MutS free from nucleotides (S10–S20) has the highest affinity for +T DNA (Kd = 20 nM; Figure 6B), and MutS bound by one (S1ATPγS–S20) or two ATPγS molecules (S1ATPγS–S2ATPγS) has the lowest affinity for +T DNA (Kd ≈ 200 and 300 nM, respectively, and only 50% binding detectable). Interestingly, at higher protein or DNA concentrations (1 µM), S1ATPγS–S20 undergoes stable interaction with +T DNA, but the S1ATPγS–S2ATPγS-+T DNA interaction remains weak (Figure 6C), similar to that observed with fully matched DNA (Figure 5A). MutS in S1ADP–S2ADP form also binds DNA with relatively high affinity (Kd = 45 nM), and most interestingly, S1ATPγS–S2ADP also binds +T DNA stably and with higher affinity (Kd = 100 nM) than S1ATPγS–S2ATPγS. If we assume that ATPγS is an appropriate mimic of ATP in this particular case, the results suggest that the S1ATP–S2ADP species—formed specifically in the presence of mismatched DNA—can rebind the mismatch (note that no interaction is detectable between S1ATPγS–S2ADP and fully matched DNA, data not shown). A recent report from the Modrich research group also indicates that human Msh2–Msh6 in the presence of both ATPγS (or AMPPNP) and ADP binds mismatched DNA with intermediate affinity relative to Msh2–Msh6 in the absence of nucleotides or in the presence of high concentrations of ATPγS (or AMPPNP) (47).
The rates of dissociation of +T DNA from MutS support the possibility that MutS can release and rebind the mismatch during its ATPase cycle. +T DNA dissociates from MutS bound to two ATP with a 10-fold faster rate constant (0.077 s−1) than from MutS free of nucleotide (0.007 s−1) (Figure 6D; note that because there were no blocks at the ends of linear +T DNA, the measurement reflects MutS dissociation from the mismatch, MutS slipping off the DNA ends, or both, ref 21). This faster dissociation rate is still comparable to the rate of formation of S1ATP–S2ADP in the ATPase pathway at 0.2–0.3 s−1 (Figure 5C and Figure 6A). Consistent with our results, the rate constants for mismatched DNA dissociation from E. coli MutS in the presence of ATP range from 0.01 to 0.02 s−1 and are slower than or comparable to the 0.06 s−1 rate constant measured for ATPase activity (21). Thus, we speculate that after ATP binds to the MutS•+T DNA complex, the protein can release the mismatch, but ATP hydrolysis at the S2 subunit allows it to rebind the mismatch (Figure 7).
It is not clear to us yet what role mismatch binding and release cycles might play in the DNA repair process. Dynamic MutS–DNA interactions may be part of a kinetic proofreading mechanism by which MutS distinguishes mismatches from correct DNA before signaling initiation of DNA repair. The cycling may also constitute a mechanism for recruitment of other repair proteins to the mismatch. Prior studies have shown that ternary complexes of MutS, MutL, and mismatched DNA are formed in the presence of ATPγS (27) or ADP•beryllium fluoride (which does not support T. aquaticus MutS release from the mismatch, ref 49), suggesting that ATP hydrolysis, MutS movement on DNA, or both may not be necessary for recruitment of MutL to the mismatch. However, absent analysis of ternary complex formation with MutS in the S1ATP–S2ADP form, we cannot rule out the possibility that this species facilitates recruitment of MutL and possibly other proteins to the mismatch and coupling of mismatch recognition to initiation of DNA repair.
Only one of the various nucleotide-bound MutS species detected thus far exhibits significant loss of affinity for the mismatch (S1ATP–S2ATP, Figure 6C,D). The transient nature of this low-affinity state during the ATPase reaction implies that MutS may not dissociate and move away from the mismatch for a significant length of time. This idea is supported by a recent report suggesting that MutS can stimulate initiation of excision on DNA substrates containing blocks between the mismatch and excision sites (26); that is, MutS may not translocate the full distance between the mismatch and excision sites to signal initiation of DNA repair, and communication between MutS and proteins working at the excision site could occur via higher-order complex formation at the mismatch itself.
Key Questions Remain
Ultimately, analysis of the different nucleotide-bound forms of MutS in the context of a complete DNA repair reaction is necessary to understand fully their role in the repair pathway. In the meantime, proteins such as T. aquaticus MutS that form a variety of well-defined and fairly stable nucleotide-bound species can serve as useful tools for investigating how events in the MutS ATPase pathway are coupled to its interactions with DNA as well as other repair proteins. Analysis of eukaryotic MutS heterodimers can help resolve other important issues, such as the role of each subunit in mismatch recognition and repair. Steady-state measurements of mutant and wild-type Msh2–Msh6 mixed heterodimers indicate that the ATPase rate is more severely impacted by mutations in Msh6 Walker motifs than by identical mutations in Msh2; only Msh6 has the mismatch-binding phenylalanine residue (18, 32, 33). While these data facilitate important hypotheses regarding Msh2 and Msh6 function, we would argue again that it is impossible to resolve conclusively the contribution of each subunit to the ATPase activity when the experiments measure only the posthydrolytic rate-limiting step or steps in the reaction. We anticipate that combining a rapid kinetic approach with new and previously developed clever biochemical experiments (such as the mixed heterodimers noted above) will help answer ever more complex questions regarding the mechanism of MutS action in DNA mismatch repair.
ACKNOWLEDGMENT
We thank Dr. Wei Yang and Dr. Peggy Hsieh for the gift of the overexpression clone of Thermus aquaticus MutS protein and for stimulating discussions. We also thank Dr. Smita Patel and Dr. Linda Bloom for advice on Pi release experiments and other helpful conversations.
Footnotes
This work was supported by a grant from the N.I.H. (No. GM 64514-01).
Abbreviations: Msh, MutS homolog; ATP, adenosine 5′ triphosphate; ATPγS, adenosine 5′-O-(3′-thiotriphosphate); ADP, adenosine 5′ diphosphate; MDCC, N-[2(1-maleimidyl)ethyl]-7-(diethylamino)-coumarin-3-carboxamide; PBP, phosphate binding protein.
REFERENCES
- 1.Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 2.Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 3.Hall MC, Matson SW. The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J. Biol. Chem. 1999;274:1306–1312. doi: 10.1074/jbc.274.3.1306. [DOI] [PubMed] [Google Scholar]
- 4.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 5.Jiricny J. Replication errors: cha(lle)nging the genome. EMBO J. 1998;17:6427–6436. doi: 10.1093/emboj/17.22.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr. Opin. Genet. Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 7.Buermeyer AB, Deschenes SM, Baker SM, Liskay RM. Mammalian DNA mismatch repair. Annu. Rev. Genet. 1999;33:533–564. doi: 10.1146/annurev.genet.33.1.533. [DOI] [PubMed] [Google Scholar]
- 8.Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 2000;34:359–399. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 9.Bellacosa A. Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ. 2001;8:1076–1092. doi: 10.1038/sj.cdd.4400948. [DOI] [PubMed] [Google Scholar]
- 10.Haber LT, Pang PP, Sobell DI, Mankovich JA, Walker GC. Nucleotide sequence of the Salmonella typhimurium mutS gene required for mismatch repair: homology of MutS and HexA of Streptococcus pneumoniae. J. Bacteriol. 1988;170:197–202. doi: 10.1128/jb.170.1.197-202.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haber LT, Walker GC. Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding activities. EMBO J. 1991;10:2707–2715. doi: 10.1002/j.1460-2075.1991.tb07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopfner KP, Tainer JA. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol. 2003;13:249–255. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 13.Junop MS, Obmolova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol, Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 14.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 15.Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK. The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 16.Kato R, Kataoka M, Kamikubo H, Kuramitsu S. Direct observation of three conformations of MutS protein regulated by adenine nucleotides. J. Mol. Biol. 2001;309:227–238. doi: 10.1006/jmbi.2001.4752. [DOI] [PubMed] [Google Scholar]
- 17.Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol. Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 18.Studamire B, Quach T, Alani E. Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol. Cell. Biol. 1998;18:7590–7601. doi: 10.1128/mcb.18.12.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joshi A, Sen S, Rao BJ. ATP-hydrolysis-dependent conformational switch modulates the stability of MutS-mismatch complexes. Nucleic Acids Res. 2000;28:853–861. doi: 10.1093/nar/28.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hess MT, Gupta RD, Kolodner RD. Dominant Saccharomyces cerevisiae msh6 mutations cause increased mispair binding and decreased dissociation from mispairs by Msh2-Msh6 in the presence of ATP. J. Biol. Chem. 2002;277:25545–25553. doi: 10.1074/jbc.M202282200. [DOI] [PubMed] [Google Scholar]
- 21.Selmane T, Schofield MJ, Nayak S, Du C, Hsieh P. Formation of a DNA mismatch repair complex mediated by ATP. J. Mol. Biol. 2003;334:949–965. doi: 10.1016/j.jmb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Blackwell LJ, Martik D, Bjornson KP, Bjornson ES, Modrich P. Nucleotide-promoted release of hMutSα from heteroduplex DNA is consistent with an ATP-dependent translocation mechanism. J. Biol. Chem. 1998;273:32055–32062. doi: 10.1074/jbc.273.48.32055. [DOI] [PubMed] [Google Scholar]
- 23.Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 25.Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol. Cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Hays JB. Signaling from DNA mispairs to mismatch-repair excision sites despite intervening blockades. EMBO J. 2004;23:2126–2133. doi: 10.1038/sj.emboj.7600153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Habraken Y, Sung P, Prakash L, Prakash S. ATP-dependent assembly of a ternary complex consisting of a DNA mismatch and the yeast MSH2-MSH6 and MLH1-PMS1 protein complexes. J. Biol. Chem. 1998;273:9837–9841. doi: 10.1074/jbc.273.16.9837. [DOI] [PubMed] [Google Scholar]
- 28.Schofield MJ, Nayak S, Scott TH, Du C, Hsieh P. Interaction of Escherichia coli MutS and MutL at a DNA mismatch. J. Biol. Chem. 2001;276:28291–28299. doi: 10.1074/jbc.M103148200. [DOI] [PubMed] [Google Scholar]
- 29.Bjornson KP, Modrich P. Differential and simultaneous adenosine di- and triphosphate binding by MutS. J. Biol. Chem. 2003;278:18557–18562. doi: 10.1074/jbc.M301101200. [DOI] [PubMed] [Google Scholar]
- 30.Lamers MH, Winterwerp HH, Sixma TK. The alternating ATPase domains of MutS control DNA mismatch repair. EMBO J. 2003;22:746–756. doi: 10.1093/emboj/cdg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antony E, Hingorani MM. Mismatch recognition-coupled stabilization of Msh2-Msh6 in an ATP-bound state at the initiation of DNA repair. Biochemistry. 2003;42:7682–7693. doi: 10.1021/bi034602h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drotschmann K, Yang W, Kunkel TA. Evidence for sequential action of two ATPase active sites in yeast Msh2-Msh6. DNA Repair. 2002;1:743–753. doi: 10.1016/s1568-7864(02)00081-2. [DOI] [PubMed] [Google Scholar]
- 33.Iaccarino I, Marra G, Palombo F, Jiricny J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSα. EMBO J. 1998;17:2677–2686. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bjornson KP, Allen DJ, Modrich P. Modulation of MutS ATP hydrolysis by DNA cofactors. Biochemistry. 2000;39:3176–3183. doi: 10.1021/bi992286u. [DOI] [PubMed] [Google Scholar]
- 35.Bowers J, Sokolsky T, Quach T, Alani E. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2-MSH6 complex disrupts mismatch recognition. J. Biol. Chem. 1999;274:16115–16125. doi: 10.1074/jbc.274.23.16115. [DOI] [PubMed] [Google Scholar]
- 36.Dufner P, Marra G, Raschle M, Jiricny J. Mismatch recognition and DNA-dependent stimulation of the ATPase activity of hMutSα is abolished by a single mutation in the hMSH6 subunit. J. Biol. Chem. 2000;275:36550–36555. doi: 10.1074/jbc.M005987200. [DOI] [PubMed] [Google Scholar]
- 37.Biswas I, Ban C, Fleming KG, Qin J, Lary JW, Yphantis DA, Yang W, Hsieh P. ligomerization of a MutS mismatch repair protein from Thermus aquaticus. J. Biol. Chem. 1999;274:23673–23678. doi: 10.1074/jbc.274.33.23673. [DOI] [PubMed] [Google Scholar]
- 38.Brune M, Hunter JL, Corrie JE, Webb MR. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 39.Jeong YJ, Kim DE, Patel SS. Kinetic pathway of dTTP hydrolysis by hexameric T7 helicase-primase in the absence of DNA. J. Biol. Chem. 2002;277:43778–43784. doi: 10.1074/jbc.M208634200. [DOI] [PubMed] [Google Scholar]
- 40.Baird CL, Gordon MS, Andrenyak DM, Marecek JF, Lindsley JE. The ATPase reaction cycle of yeast DNA topoisomerase II. Slow rates of ATP resynthesis and P(i) release. J. Biol. Chem. 2001;276:27893–27898. doi: 10.1074/jbc.M102544200. [DOI] [PubMed] [Google Scholar]
- 41.Bertram JG, Bloom LB, Hingorani MM, Beechem JM, O’Donnell M, Goodman MF. Molecular mechanism and energetics of clamp assembly in Escherichia coli. The role of ATP hydrolysis when γ complex loads β on DNA. J. Biol. Chem. 2000;275:28413–28420. doi: 10.1074/jbc.M910441199. [DOI] [PubMed] [Google Scholar]
- 42.Biswas I, Hsieh P. Identification and characterization of a thermostable MutS homolog from Thermus aquaticus. J. Biol. Chem. 1996;271:5040–5048. doi: 10.1074/jbc.271.9.5040. [DOI] [PubMed] [Google Scholar]
- 43.Brune M, Hunter JL, Howell SA, Martin SR, Hazlett TL, Corrie JE, Webb MR. Mechanism of inorganic phosphate interaction with phosphate binding protein from Escherichia coli. Biochemistry. 1998;37:10370–10380. doi: 10.1021/bi9804277. [DOI] [PubMed] [Google Scholar]
- 44.Marsischky GT, Kolodner RD. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J. Biol. Chem. 1999;274:26668–26682. doi: 10.1074/jbc.274.38.26668. [DOI] [PubMed] [Google Scholar]
- 45.Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J. Biol. Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 46.Wang H, Yang Y, Schofield MJ, Du C, Fridman Y, Lee SD, Larson ED, Drummond JT, Alani E, Hsieh P, Erie DA. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martik D, Baitinger C, Modrich P. Differential specificities and simultaneous occupancy of human MutSα nucleotide binding sites. J. Biol. Chem. 2004;279:28402–28410. doi: 10.1074/jbc.M312108200. [DOI] [PubMed] [Google Scholar]
- 48.Gradia S, Acharya S, Fishel R. The role of mismatched nucleotides in activating the hMSH2-hMSH6 molecular switch. J. Biol. Chem. 2000;275:3922–3930. doi: 10.1074/jbc.275.6.3922. [DOI] [PubMed] [Google Scholar]
- 49.Alani E, Lee JY, Schofield MJ, Kijas AW, Hsieh P, Yang W. Crystal structure and biochemical analysis of the MutS.ADP.beryllium fluoride complex suggests a conserved mechanism for ATP interactions in mismatch repair. J. Biol. Chem. 2003;278:16088–16094. doi: 10.1074/jbc.M213193200. [DOI] [PubMed] [Google Scholar]