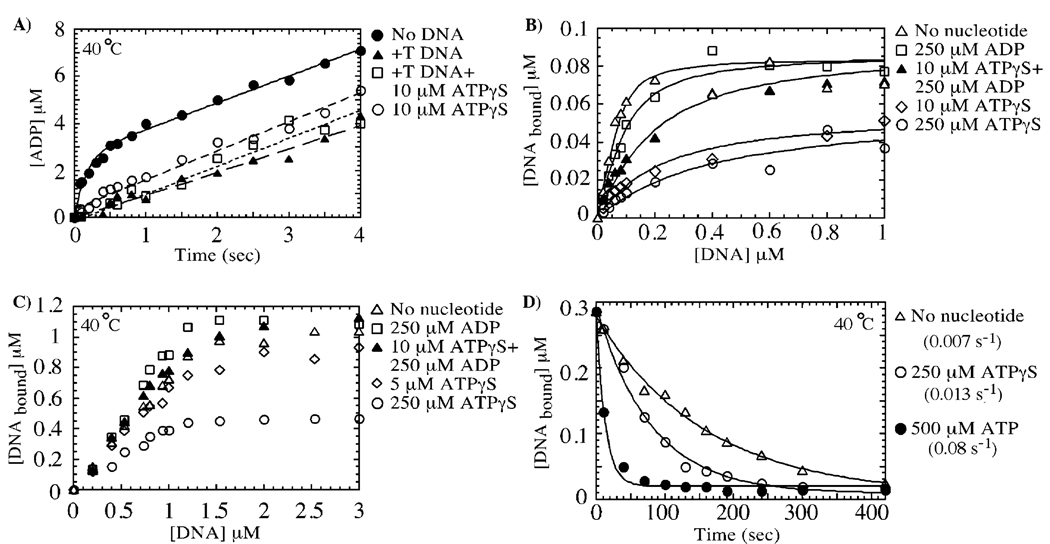
FIGURE 6.
Mismatched DNA inhibits ATP hydrolysis specifically at the high-affinity site, yielding an ATP/ADP-bound form of MutS, which has high affinity for mismatched DNA. In panel A, +T DNA binding to MutS (3 µM dimer) suppresses the burst of ATP hydrolysis (▲), but the slow phase remains identical to that in the absence of DNA (●, 0.33 s−1). The same slow ATPase rate is observed for MutS preincubated with 10 µM ATPγS (○, high-affinity site occupied, no burst phase) or with 10 µM ATPγS and +T DNA (□), indicating that the low-affinity site continues to hydrolyze ATP when MutS is bound to mismatched DNA. Panels B and C show nitrocellulose membrane filtration experiments of 0.1 and 1 µM MutS dimer, respectively, binding to +T DNA in the absence of nucleotide cofactors (∆, Kd = 20 nM), in the presence of 250 µM ADP (□, both high-affinity and low-affinity sites occupied, Kd = 45 nM), and in the presence of 10 µM ATPγS and 250 µM ADP (▲, ATPγS in high-affinity site and ADP in low-affinity site, Kd = 100 nM). The interaction between MutS and +T DNA is weaker in the presence of 10 µM ATPγS (◊, one site occupied by ATPγS, Kd = 200 nM; note stable +T DNA interaction with this species at 1 µM MutS) and 250 µM ATPγS (○, both sites occupied with ATPγS, Kd ≈ 300 nM; note weak +T DNA interaction with this species even at 1 µM MutS). In panel D, the rate of dissociation of 32P-labeled +T DNA (1 µM) from MutS (0.5 µM) was measured over time by membrane filtration assays using 50 µM unlabeled +T DNA chase (with or without nucleotides). The dissociation rate is 0.007 s−1 in the absence of nucleotides (∆), 0.013 s−1 in the presence of 250 µM ATPγS (○), and 0.077 s−1 in the presence of 500 µM ATP (●).