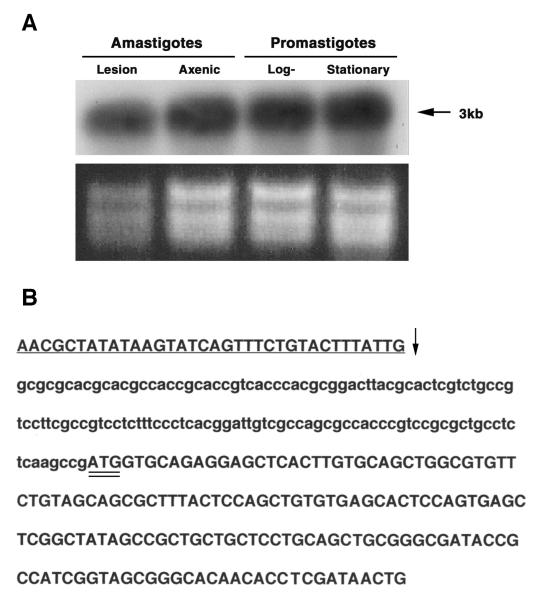
FIG. 3. Northern analyses and mapping of the spliced-leader acceptor site.
A. Northern blot analysis of LmexCht1 mRNA transcripts present in various L. mexicana parasite developmental forms. Top Panel. Total RNA (5 μg) isolated from L. mexicana mouse lesion amastigotes (i.e. in vivo-derived), axenic amastigotes (i.e. in vitro-grown), and log (procyclic)- and stationary (metacyclic)-phase promastigotes was separated in an agarose gel, transferred onto a nylon membrane and hybridized with the LmexCht1-DIG 270 probe (i.e. corresponding to nt 502– nt 771 of the LmexCht1-ORF). Arrow indicates the position of the ~3 kb LmexCht1 mRNA transcript in these samples. Bottom Panel. Ethidium bromide strained agarose gel (used in panel A) showing the ribosomal RNA in each of the total RNA samples used above. B. Mapping of the LmexCht1 5′-spliced-leader acceptor site. Nucleotide sequence of the RT-PCR product obtained with LmexCht1 mRNA amplified with an L. mexicana-spliced leader (forward: SpliceFwd) oligonucleotide primer and an internal LmexCht1 (reverse: ORF-RT/Rev) primer. The 5′-untranslated region of the LmexCht1 gene is shown in lower case letters. The LmexCht1 open reading frame is shown in Upper case letters, a portion of the conserved spliced leader sequence (i.e. forward primer) is shown in underlined Caps and the arrow (↓) marks the position of the spliced-leader acceptor site. The ATG-start codon of the LmexCht1 open reading frame is double-underlined.