Figure 4.
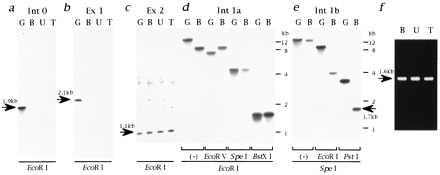
Cloning of the genomic DNA containing the deletion breakpoint of the CM hamsters. Genomic DNAs digested with EcoRI were hybridized by the Int 0 (a), Ex 1 (b), or Ex 2 (c) probes. The sizes of hybridized bands are shown by arrows. (d) Genomic DNAs digested with EcoRI followed by EcoRV, SpeI, or BstXI were hybridized by the Int 1a probe. Note that restriction fragment length polymorphisms were detected for EcoRI/EcoRV but not for EcoRI/SpeI digestion. (e) Genomic DNAs digested with SpeI followed by EcoRI or PstI were hybridized by the Int 1b probe. Note that the 1.7-kb polymorphic band (shown by an arrow) was detected for SpeI/PstI digestion. The same results as d and e were obtained for both U and T (data not shown). (f) Amplification of polymorphic PstI–SpeI fragments of the CM hamster genomes. The size of the amplified fragment (1.6 kb; shown by an arrow) is smaller than that of the SpeI–PstI restriction fragment length polymorphism band (1.7 kb), because the GS primer used for this cloning is located 80 bp 5′ upstream from the SpeI cleavage site (Fig. 3 Bottom).
