Figure 6.
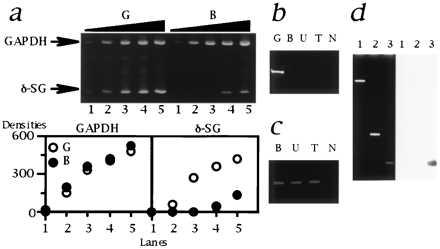
Analysis of δ-SG transcript in the CM hamsters. (a) Multiplex RT-PCR. (Upper) The sizes of PCR products for GAPDH and δ-SG (indicated by arrows) are 452 and 150 bp, respectively. Lanes: 1, 2/1 μl; 2, 2/4 μl; 3, 2/16 μl; 4, 2/64 μl; 5, 2/256 μl of template cDNA. The forward (exIIF) and reverse (exIIR2) primers were both located in the second exons. (Lower) The densities of each band quantified by nih image software are plotted. Note that GAPDH, an internal standard, was equivalently amplified. The amount of δ-SG transcript in B (•) was estimated to be 20–40 times lower than that of G (○). The similar data were also obtained for U and T (data not shown). (b) RT-PCR targeted for the authentic first and the second exons of δ-SG. The forward (exIF) and reverse (exIIR1) primers were located in the authentic first and the second exons, respectively. Note that a 217-bp band was detected only for G. N, no template. (c) 5′ RACE. The size of the bands was 0.5 kb. N, no template. (d) Localization of the alternative first exon of δ-SG. (Left) Ethidium bromide staning. (Right) Southern blot analysis with an alternative first exon probe (AlEx 1; 274-bp PCR products amplified with a primer set of alexIF and alexIR). Lanes: 1, 13.3-kb EcoRI–EcoRI fragment of the first intron of normal δ-SG gene containing the deletion breakpoint (Fig. 3 Middle); 2, 1.1-kb Ex 2 fragment (Fig. 3 Upper); 3, AlEx 1 probe as a positive control for hybridization. Note that AlEx 1 probe did not hybridize the genomic fragments of normal hamsters corresponding to lanes 1 and 2 that cover the genomic region between the deletion breakpoint and the second exon of δ-SG of the CM hamsters.
