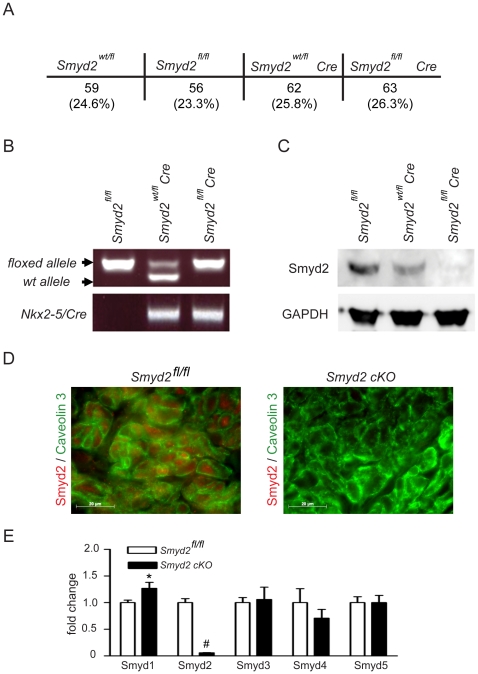
Figure 4. Analysis of cardiac specific Smyd2 deletion.
(A) Mating of Smyd2fl/fl mice with Smyd2fl/flCre mice resulted in offspring of four genotypes (Smyd2wt/fl, Smyd2fl/fl, Smyd2wt/flCre, Smyd2fl/flCre) at normal Mendelian ratios. (B) The genotype of the animals used for analysis is shown by a representative genotyping PCR. Animals homozygous for the floxed Smyd2 allele but lacking the Nkx2–5 driven Cre recombinase were used as control. (C) Protein extracts (70 µg) from Smyd2fl/fl, Smyd2wt/flCre or Smyd2fl/flCre (cKO) mouse hearts were subjected to western-blot analysis and the knockdown efficiency as well as antibody specificity was assessed by probing the blot with an anti-Smyd2 antibody. Smyd2 protein expression was lowered by half in the heterozygous animals while it was almost completely absent in the homozygous cKO animals. The blot was re-probed with an anti-GAPDH antibody for equal loading control. (D) Cryosections from P1 control or Smyd2 cKO mice were stained with an anti-Smyd2 antibody (red) and an anti-Caveolin-3 antibody (green) to co-stain the cardiomyocyte cell membrane. Smyd2 shows a distinct expression in the cardiomyocytes of control mice while no expression was observed in the cardiomyocytes of cKO animals. Pictures were taken at a magnification of 1000×. (E) Real-time qPCR showing relative expression levels of Smyd-family members 1–5 in P5 control or Smyd2 cKO mouse heart ventricles. Data is shown as mean +/− SEM, *p<0.05 vs. control, #p<0.01 vs. control, n = 5.