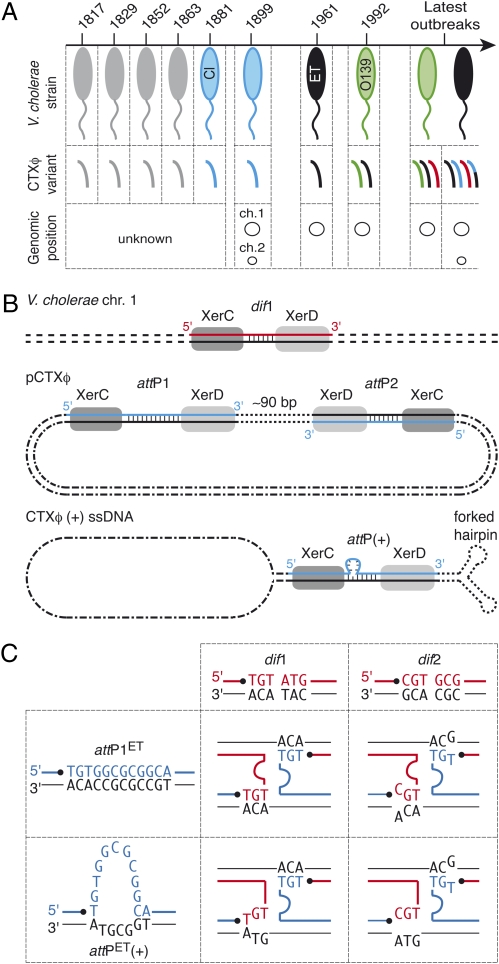
Fig. 1.
Lysogenic conversion of V. cholerae strains by CTXφ. (A) CTXφ variants found in cholera epidemic strains. V. cholerae strains: gray, unknown; blue, classical; black, El Tor; green, O139. CTXφ variants: gray, unknown; blue, classical; black, El Tor; green, O139; red, G; black and blue, El Tor and classical hybrid. (B) Scheme of Xer recombination sites. dif1, dimer resolution site of the N16961 chromosome 1; attP1 and attP2, Xer sites found in the replicative form of CTXφ (pCTXφ); attP(+), the recombination site created by the folding of the (+) ssDNA genome of CTXφ. The two DNA strands of each site are drawn. The strand cleaved by XerC is shown in color. Vertical bars indicate bases present in the overlap region of each site. (C) Schemes of the Watson–Crick bp interactions that could stabilize the strand exchange catalyzed by XerC between the overlap regions of the two chromosome dimer resolution sites of N16961 and the two putative attachment sites of El Tor variants of CTXφ. dif1, chromosome 1 dimer resolution site; dif2, chromosome 2 dimer resolution site; attP1ET, attP1 found in CTXφ El Tor variants; attP(+)ET, attachment site unmasked by the folding of the (+) ssDNA genome of El Tor variants of CTXφ. The strands cleaved by XerC on dimer resolution and phage attachment sites are shown in red and in blue, respectively. Pairing interactions are indicated by the proximity of the bases.