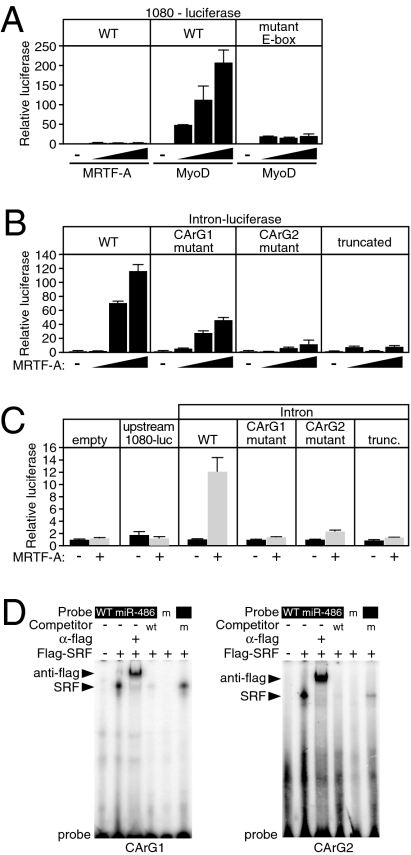
Fig. 3.
Identification of miR-486/sAnk1 regulatory sequences in vitro. (A) Luciferase assay demonstrating the response of 1080 bp of upstream DNA (WT) or of the DNA fragment containing a mutation of the E-boxes to increasing amounts of MRTF-A and MyoD. A total of 50, 100, and 200 ng of expression plasmids were cotransfected with the luciferase reporter construct in COS cells. Error bars represent SD. (B) Response of the 649 bp of sAnk1 intron 39a to MRTF-A examined by luciferase reporter assay in COS cells. A total of 50, 100, and 200 ng of expression plasmid were transfected with full-length intron–luciferase (WT), a luciferase reporter construct with a mutation in the proximal CArG1, a mutation in the distal CArG2, or a truncation that consists of the 5′ most 300 bp. Error bars represent the SD. (C) Response of the empty luciferase vector or the 1,080-bp upstream luciferase or various intron–luciferase reporters to adenovirus-mediated MRTF-A expression in CMCs. Error bars represent SD. (D) Gel electromobility shift assay demonstrating the binding of flag epitope-tagged SRF overexpressing COS cell lysate to a radiolabeled oligonucleotide probe consisting of the sequences of the CArG1 or CArG2 of sAnk1 intron 39a. Flag antibody results in a supershift, and wild-type unlabeled competitor abolishes the binding of CArG probe. Mutant probe does not bind SRF, nor does mutant unlabeled competitor abolish WT probe binding to SRF. m, mutant CArG.