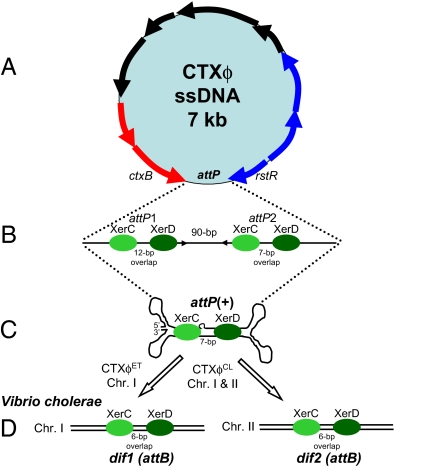
Fig. 1.
(A) Schematic representation of the single-stranded CTXϕ genome. Arrows indicate ORFs identified and characterized in CTXϕ. Phage attachment site is indicated by attP. (B) Linear representation of ssDNA attP site and regions identified by Huber and Waldor (9) and by McLeod and Waldor (5) as essential for efficient integration of CTXϕ into chromosome I (ChrI) of V. cholerae. Green oval shapes represent XerC and XerD attached at their respective binding sites on attP sites 1 (attP1) and 2 (attP2), identified by McLeod and Waldor (5). Also shown are the spacer or overlap regions between XerC and XerD binding sites, 12 bp for attP1 and 7 bp for attP2. (C) Secondary fork and stem structure formed when attP1 and attP2 base pair, creating the new attP+ site, identified by Val et al. (6). (D) The bacterial attachment sites (attB), dif1 and dif2 on chromosome I and chromosome II, the primary function of which is to resolve chromosomal dimers during cell division.