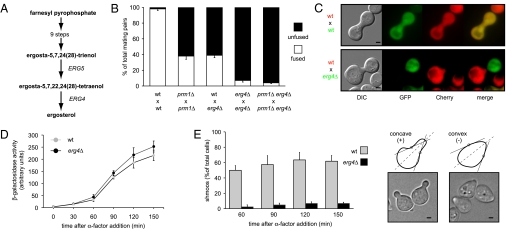
Fig. 1.
ERG4 mutant is defective in cell fusion. (A) Schematic representation of the ergosterol biosynthetic pathway highlighting the last two steps. (B) Quantitative cell fusion assays. All phenotypes were tested for both mating types, and the results were indistinguishable. Error bars indicate standard errors. (C) erg4Δ cells form morphologically abnormal mating pairs. MATα cells carrying cytoplasmic GFP were mixed with MATa cells carrying Pgk1-mCherry on nitrocellulose filters and incubated on YPD plates for 3 h at 30 °C. Fixed mating mixtures were then imaged by DIC and wide-field fluorescence microscopy. (Scale bars: 2 μm.) (D) erg4Δ cells preserve a normal pheromone response pathway. Strains harboring a FUS1-LACZ transcriptional reporter were grown and treated with 6 μM α-factor. At different time points, cells were harvested and assayed for β-galactosidase activity. Error bars indicate SEs. (E) erg4Δ cells are defective for shmoo formation. MATa cells were treated with α-factor, and aliquots were collected, fixed, and observed by bright-field microscopy at the indicated time points. For quantification (Left), cells that showed two convex edges along the axis of polarization (dotted lines) were scored as positive hits (Right). (Scale bars: 1 μm.)