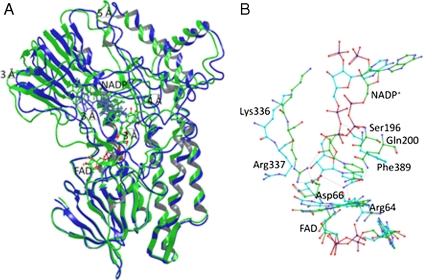
Fig. 3.
Cartoon representation of the superimposed WT PAMO (Green) and mutant Gln93Asn/Pro94Asp (Blue) structures. (A) Average structures were obtained from the conformers of 1 ns of MD trajectory after 5 ns of equilibration. Productive MD simulations were run during 6 ns including coenzyme FAD and NADP+. Structural superposition was performed using FAD molecule and FAD-binding residues (residues within 3 Å) as anchor. It is possible to observe important backbone rotation especially in the NADP domain. (B) Details of the NADP+ binding, comparing WT PAMO (Green) and mutant Gln93Asn/Pro94Asp (Blue). Arg337 in the mutant shifts away from the flavin ring and does not show the H-bond present in the WT.