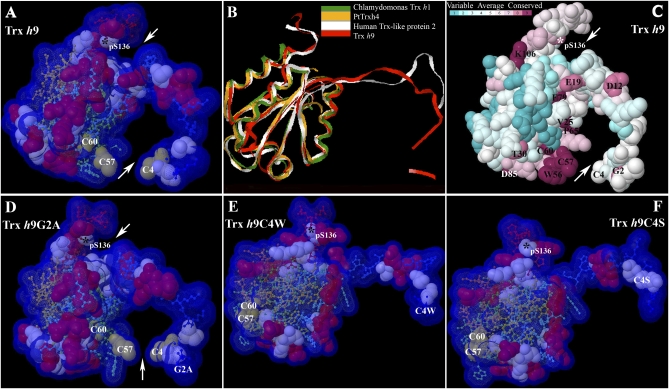
Fig. 5.
Three-dimensional models and conserved residue prediction for Trx h9. (A) Three-dimensional model of Trx h9. (B) Superimposition of 3D model of Trx h9 (red) and the top three templates of Trx h9: C. reinhardtii Trx h1 (green; PDB ID code 1ep7A); poplar PtTrxh4 (yellow; PDB ID code 3d21); and human Trx-like protein 2 (white; PDB ID code 2diyA), using 3d-SS (3-Dimensional Structural Superposition) service. (C) Conserved residue analysis of Trx h9. Residue conservation from variable to conserved is shown in green to dark red, respectively. (D–F) Three-dimensional model of Trx h9G2A, Trx h9C4W, and Trx h9C4S, respectively. Arrows point to potential docking sites of Trx h9 in A, C, and D. Cys, Ser, and positively charged residues are shown in off-yellow, light blue, and red, respectively, as 100% of van der Waals. The remainder of residues from N to C terminus are shown in blue to red as 20% of van der Waals in A and D, and E and F. All atoms are coupled with Solvent-Accessible Surface (VDW + 1.4 Å) in A and D–F. Asterisks indicate phosphorylated Ser at position 136 (pS136) at the C terminus of Trx h9. Conserved amino acids with single-letter abbreviations are indicated at their numbered position in C.