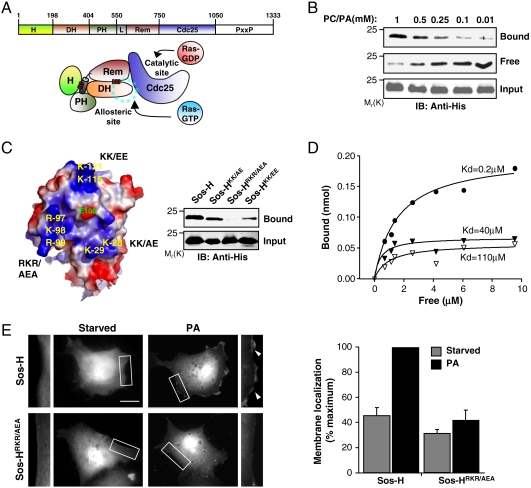
Fig. 1.
The histone domain of Sos (Sos-H) binds to membrane lipids. (A) The domain arrangement of Sos. H, Histone; DH, Dbl homology; PH, Pleckstrin homology; L, helical Linker; REM, Ras exchange motif; CDC25, yeast CDC25 homology; and PxxP, proline-rich region. Below, a cartoon depiction of the relative orientation of the various domains of Sos in the autoinhibited state. The red bars indicate intramolecular interactions. The blue dotted line circle indicates the binding site for allosteric Ras, which is occluded by the DH domain. (B) His-tagged Sos-H (0.5 μM) was mixed with the indicated concentrations of lipid vesicles comprised PC∶PA (90∶10). Vesicles were pelleted and the amount of Sos-H in the pellet (Bound) or in the supernatant (Free) was detected by immunoblotting (IB). (C) Surface electrostatic potential map (blue, positive; red, negative) showing the 3 basic patches in Sos-H and the residues mutated (see Materials and Methods). Right, Sos-H constructs containing the indicated mutations were mixed with 1 mM PC∶PA vesicles (90∶10) and binding was determined as in B. (D) Increasing amounts of Sos-H were incubated with 1 mM PC∶PA (closed circles) or PC lipid vesicles (open triangles) and Sos-HRKR/AEA with PC∶PA vesicles (closed triangles). Kd was determined as previously described (2). Data presented in panels B–D are representative of three independent experiments. (E) COS-1 cells were transfected with GFP-tagged Sos-H and membrane localization scored in either starved cells or upon incubation with PA (100 μM) for 20 min. The image represents a single slice of 0.25 μm from a serial Z-section of the cell. The boxed areas are enlarged in the left and right adjacent images with arrowheads showing the relative increase in fluorescence intensity at the membrane. Results (bar graph) are mean ± s.d. of three independent experiments with at least 100 expressing cells counted for each condition (see Materials and Methods). Scale bar: 10 μm.