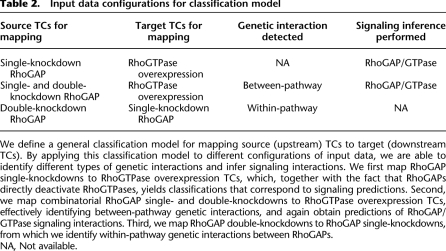
Table 2.
Input data configurations for classification model
We define a general classification model for mapping source (upstream) TCs to target (downstream TCs). By applying this classification model to different configurations of input data, we are able to identify different types of genetic interactions and infer signaling interactions. We first map RhoGAP single-knockdowns to RhoGTPase overexpression TCs, which, together with the fact that RhoGAPs directly deactivate RhoGTPases, yields classifications that correspond to signaling predictions. Second, we map combinatorial RhoGAP single- and double-knockdowns to RhoGTPase overexpression TCs, effectively identifying between-pathway genetic interactions, and again obtain predictions of RhoGAP/GTPase signaling interactions. Third, we map RhoGAP double-knockdowns to RhoGAP single-knockdowns, from which we identify within-pathway genetic interactions between RhoGAPs.
NA, Not available.