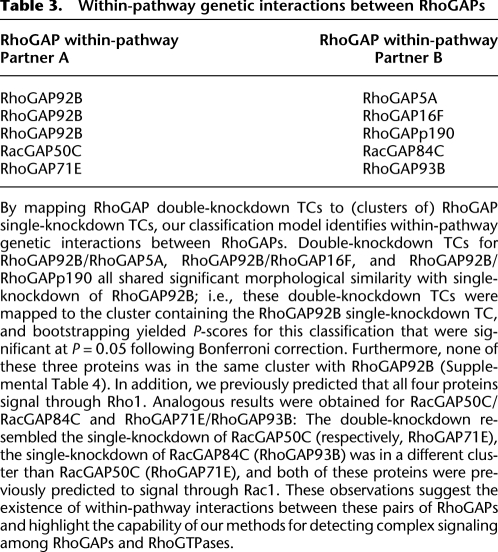
Table 3.
Within-pathway genetic interactions between RhoGAPs
By mapping RhoGAP double-knockdown TCs to (clusters of) RhoGAP single-knockdown TCs, our classification model identifies within-pathway genetic interactions between RhoGAPs. Double-knockdown TCs for RhoGAP92B/RhoGAP5A, RhoGAP92B/RhoGAP16F, and RhoGAP92B/RhoGAPp190 all shared significant morphological similarity with single-knockdown of RhoGAP92B; i.e., these double-knockdown TCs were mapped to the cluster containing the RhoGAP92B single-knockdown TC, and bootstrapping yielded P-scores for this classification that were significant at P = 0.05 following Bonferroni correction. Furthermore, none of these three proteins was in the same cluster with RhoGAP92B (Supplemental Table 4). In addition, we previously predicted that all four proteins signal through Rho1. Analogous results were obtained for RacGAP50C/RacGAP84C and RhoGAP71E/RhoGAP93B: The double-knockdown resembled the single-knockdown of RacGAP50C (respectively, RhoGAP71E), the single-knockdown of RacGAP84C (RhoGAP93B) was in a different cluster than RacGAP50C (RhoGAP71E), and both of these proteins were previously predicted to signal through Rac1. These observations suggest the existence of within-pathway interactions between these pairs of RhoGAPs and highlight the capability of our methods for detecting complex signaling among RhoGAPs and RhoGTPases.