Fig. 1.
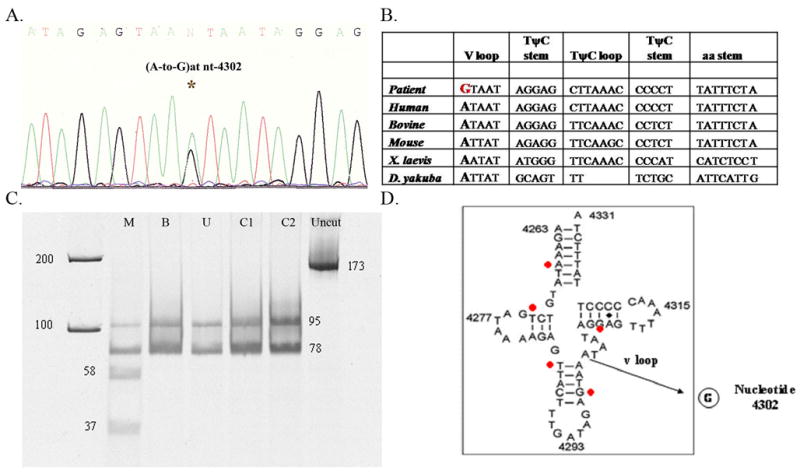
(A) Electropherogram of DNA sequence. Electrophoresed samples were analyzed with Sequence Analysis software (Perkin-Elmer Applied Biosystems). At position 4302 a heteroplasmic adenine to guanine transition is evident (4302A>G).
(B) Nucleotide sequence showing inter-species similarity of the tRNAIle gene and the highly conserved base at nt 4302. The mutation in our patient is highlighted with a bold red letter and compared with human, bovine, mouse, Xenopus (X) laevis, Drosophila (D) yakuba.
(C) PCR-RFLP analysis. Fragments after digestion with DdeI were separated on a 15% non-denaturing acrylamide gel, visualized and quantitated using SYBR Gold® Nucleic Acid Gel Stain (Invitrogen). DdeI cuts the wild type into a 95 and a 78 bp fragment, whereas mutant mtDNA was digested into three (58, 37 and 78 bp). Patient’s tissues: M, muscle; B, blood; U, urine; C1, C2 (controls); uncut PCR product.
(D) Secondary structure of the human wild type tRNAIle showing the nt. 4302 base pair substitution A-to-G at the variable loop (v loop) region. The red points show previously described mutations associated with CPEO phenotype (position 4267, 4274, 4285, 4298, and 4309).
