Fig. 3.
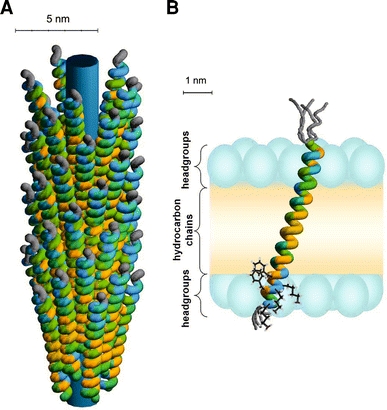
a Schematic illustration of the phage-bound model for the major coat protein of bacteriophage M13. The colour coding of the amino acid residues is based on a hydrophobicity scale (Fig. 2). Unstructured protein regions are indicated in grey. The inner cylinder indicates the viral DNA. b Structure and membrane embedding of M13 coat protein in fully hydrated vesicles of 18:1PC (and mixed phospholipid systems with C18 acyl chains), based upon recent site-directed labelling spectroscopy (Koehorst et al. 2004; Nazarov et al. 2007; Stopar et al. 2006b; Vos et al. 2005, 2007). The protein is effectively anchored with the C-terminal domain at the membrane–water interface by three “snorkelling” lysines (Lys40, Lys43, and Lys44) and two “anti-snorkelling” phenylalanines (Phe42 and Phe45). The size of the membrane regions is obtained from the literature, with the positions of the carbonyls serving as borders for the headgroup region (Ridder et al. 2002; White and Wimley 1999). The phospholipid headgroups are indicated with blue ellipsoids and the hydrocarbon chain region is coloured in yellow. The protein forms a mainly α-helical conformation tilted at 18° to the membrane normal. The first nine amino acid residues, which encompass the hydrophilic anchor (Table 1), are unstructured (as indicated by different possible gray structures). The membrane-bound protein structure does not differ much from the native α-helical structure of the protein in bacteriophage M13 in a. The protein survives the membrane-bound state by a simple tilt mechanism based on anchoring of its C-terminal domain at the membrane–water interface and a subtle structural adjustment at the extreme end of the N-terminal domain
