Fig. 2.
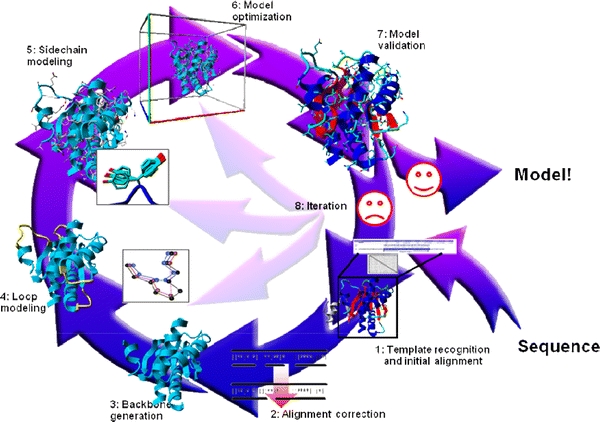
The eight steps of homology modelling. The first step involves finding a suitable homologous protein whose structure can be used as a modelling template, and generating the initial alignment between that template and the model sequence. In step 2 this alignment is refined using, for example, knowledge obtained from the template structure. In step 3 the backbone is generated and deletions are performed so that, temporarily forgetting insertions, the backbone of the template looks like that of the model as much as possible. In step 4 gaps in the model are closed, and optionally loops are constructed ab initio. In step 5, the side-chains are added using rotamer libraries to find the best rotamer for that local backbone conformation. Step 6 consists of molecular-dynamics simulation of the complete model in order to remove (the majority of) the introduced errors. In step 7 the model is checked for remaining errors using validation software. Depending on the outcome of the validation step we either approve the model or iterate the modelling process (step 8) starting from steps 1–6
