Fig. 6.
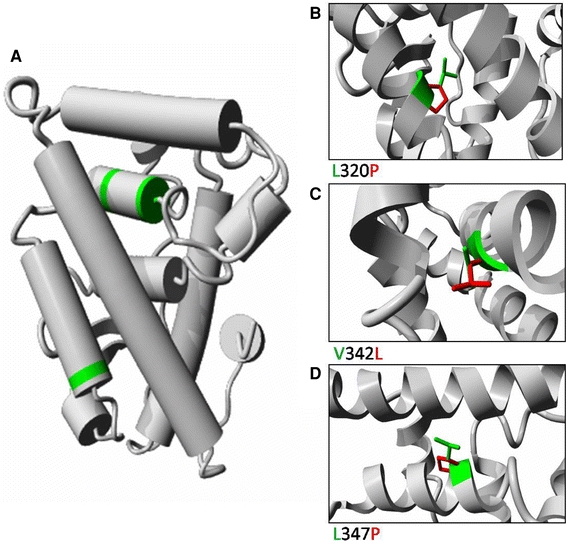
a Molecular modelling of the ligand-binding domain of the human ESRRB protein. The structure was deduced from the known ESRRB structure. The various helices are represented by cylinders. The three amino acids that are affected by the mis-sense mutations are indicated in green. Detailed views of the three mutations are shown in panels b, c and d. The wild-type residue is depicted in green, whereas the side-chain of mutant residue is presented in red. b L320 makes hydrophobic contacts in the core between two helices, the mutation to P causes loss of many hydrophobic interactions. Additionally, the P will disturb the structure of the helix. c V342 is tightly packed in the core between two helices. This core will be disturbed by the V342L mutation because L has a larger side-chain. d L347P will cause loss of hydrophobic interactions between the helices, and the introduction of P will disturb the structure of the helix
