Abstract
CREB-binding protein (CBP) is an important coactivator of basal transcription machinery and a critical regulator of cellular proliferation, differentiation, and apoptosis. It is hypothesized that CBP function is regulated by post-translational modifications, such as phosphorylation and methylation. Specific kinase-mediated phosphorylation of CBP has been shown to affect not only intrinsic histone acetyl transferase activity, but also transcriptional activity of various target promoters and interaction with binding partners. While most of the identified CBP phosphorylation sites have been mapped to the N-terminus of the protein, based on previous studies of the CBP homolog (p300), Protein Kinase B/Akt is predicted to phosphorylate the C-terminus of CBP. However, there is no direct evidence of Akt-mediated phosphorylation of CBP. Here we report the first purification procedure of recombinant fragment of CBP, encompassing the cysteine/histidine-rich domain 3 (CH3) and glutamine-rich (Q) domain of the protein, which is suitable for structural and interaction studies. We provide the first evidence of protein-protein interaction between the full-length Akt1 and the C-terminus of CBP by fluorescence spectroscopy and the subsequent phosphorylation of CBP by in vitro phosphorylation assay. Our results suggest that Akt signaling may have important implications on the in vivo molecular interaction of CBP with various transcription factors and modulation of cellular responses.
Introduction
CREB-binding protein (CBP)1 and its homolog p300 belong to a class of proteins that play a vital role in remodeling the chromatin structure at the target promoters and facilitating gene transcription by RNA polymerase. CBP and p300 serve as protein scaffolds for a number of nuclear transcription factors, thus mediating transcriptional responses to numerous intracellular and extracellular signals [1-4]. CBP and p300 regulate cell proliferation, differentiation, apoptosis, and response to viral challenges. The intrinsic histone acetyl transferase (HAT) activity of these proteins facilitates chromatin unwinding and subsequent recruitment of other coactivators to the basal transcription machinery.
While CBP and p300 share overlapping functions, significant differences exist. For instance, mice that are heterozygous for CBP gene (Crebbp+/−) show defects in hematopoietic differentiation, while Ep300+/− mice do not show any such abnormal differentiation [5]. Moreover, studies have shown that CBP and p300 associate with different proteins and have different substrate specificity [6]. Regardless of their differences, defects in CBP and p300 expression have been implicated in several human diseases, ranging from developmental disorders to cancer progression [7].
Several studies have implicated CBP as the rate-limiting cofactor in gene transcription [8-13], suggesting a model of competitive binding between transcription factors that require CBP for activation. One mechanism by which CBP activity is regulated in the cells is through post-translational modifications, such as phosphorylation, acetylation, ubiquitinylation, and methylation [14]. Thus, post-translational modifications of CBP may bring about changes in its HAT activity, protein-protein interactions, substrate specificity, or stability [14-16]. It is well established that CBP is a target of various protein kinases, including MAPK/ERK, CaMKIV, and IKKα [2, 17-25]. Several specific phosphorylation sites have been identified at the N-terminal half of the CBP [18-20]. For instance, CBP phosphorylation at S301 and S436 resulted in increased HAT and transcriptional activity during neuronal differentiation [18] and recruitment of transcription factors AP-1 and Pit-1 via growth factor signaling pathways [19], respectively. Recently, IKKα-mediated phosphorylation of CBP at S1382 and S1386 resulted in increased HAT activity as well as tumor growth and upregulation of anti-apoptotic NF-κB-responsive genes due to a switch in binding preference of CBP from p53 to NF-κB [20]. Although numerous studies have implicated the C-terminal region of CBP as a target of phosphorylation [13, 17, 21-25], only in the past few years that specific phosphorylation sites have been identified. In particular, phosphorylations of CBP Ser-2016 [25] and Ser-2080 [13] by ERK1 have been shown to be important for transcriptional activity in cardiac cells and regulation of coactivator association with AP-1 sites in the MMP-9 promoter, respectively.
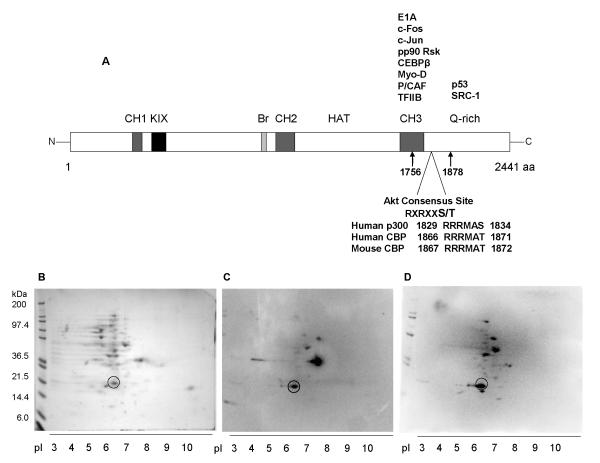
Previous studies have shown that human p300 (residues 1829-1834) contains a consensus motif for Protein kinase B/Akt wherein Ser-1834 serves as the phospho-acceptor site [26, 27]. PKB/Akt, a key player in the PI3K/Akt/mTOR signaling pathway, belongs to a family of serine/threonine kinases that regulates various cellular processes, including proliferation, cell cycle progression, metabolism, angiogenesis, motility and apoptosis [28, 29]. The Akt consensus motif is conserved in human (residues 1866-1871) and mouse (residues 1867-1872) CBP and spans a portion of the cysteine/histidine-rich domain 3 (CH3) and glutamine-rich domain (Q) of the protein [26] (Fig. 1A). Phosphorylation of p300 Ser-1834 by Akt blocks the transactivation of the CAAT-enhancer binding protein [26] and increases the transcriptional activity of ICAM-1 promoter [27]. While the corresponding residue in CBP (Thr-1871 in human and Thr-1872 in mouse) has been suggested to be an Akt phosphorylation site, there is no direct evidence for this. Given the location of the Akt consensus motif within the region of CBP that interacts with other proteins that play a role in the regulation of cellular growth and differentiation [30], it will be interesting to examine the exact contribution of PI3K/Akt/mTOR signaling to these cellular processes (Fig. 1A).
Fig. 1.
Experimental pI of T7-tagged fragment of CBP spanning the CH3 domain and partial Q-domain, CBP CH3. (A) Diagram of different domains of CBP, including partial list of known interacting proteins at the C-terminus. Location of the Akt consensus site within the cloned fragment of mouse CBP is indicated. The analogous Akt consensus sites at human CBP and p300 are also shown. (B) Two-dimensional gel electrophoresis of CBP CH3 lysates and subsequent detection of resolved proteins via Coomassie blue staining and (C) parallel Western blotting against T7 tag and (D) Akt substrate motif.
Our laboratory has shown that CBP is required for cellular growth arrest and apoptosis in acutely damaged human mammary epithelial cells (HMECs) [12, 31-33]. In particular, apoptosis induced in HMECs by the partial estrogen antagonist/agonist Tamoxifen (Tam) is associated with rapid loss of both phosphorylation at Akt Ser-473 and Akt activity [32]. In the present study, we have employed site-directed mutagenesis and in vitro interaction techniques to investigate whether the C-terminal CBP fragment spanning the Akt consensus motif is a direct binding partner and phosphorylation site of Akt. Here we demonstrate that the C-terminus of CBP can interact directly with full-length Akt1 in vitro and contains a reversible Akt-mediated phosphorylation site(s). Our in vitro data suggests a potential role of Akt1-mediated phosphorylation of CBP in modulating a wide variety of cellular responses, as proposed by other groups [24, 26, 27, 34].
Materials and methods
Materials
Bis-Tris propane, DTT, EDTA, ammonium sulfate, kanamycin, and chloramphenicol were purchased from Sigma-Aldrich (St. Louis, MO). BugBuster® protein extraction reagent, Benzonase® nuclease, bacterial expression vector pET24b, competent cells BL21(DE3) and BL21(DE3)pLysS, T7 promoter primer, T7•Tag® antibody HRP conjugate were from Novagen (Madison, WI). Protease inhibitor cocktail set II and IPTG were purchased from Calbiochem (San Diego, CA). CHT™ ceramic hydroxyapatite type I resin, gel filtration standards, Bradford assay reagent, and polyvinylidene difluoride (PVDF) membrane were from Bio-Rad (Hercules, CA). High performance Superdex 200 10/300 GL column was purchased from GE Healthcare (Piscataway, NJ). The activated GST fusion protein of full-length human Akt1 (GST-Akt1) and rabbit phospho-Akt substrate monoclonal antibody were from Cell Signaling Technology (Danvers, MA). It should be noted that activated GST-Akt1 contains T308D/S473D double mutations. Donkey polyclonal anti-rabbit IgG HRP conjugate and rabbit polyclonal IgG against the C-terminus of CBP (C-20) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
PCR mutagenesis of CH3 CBP fragment
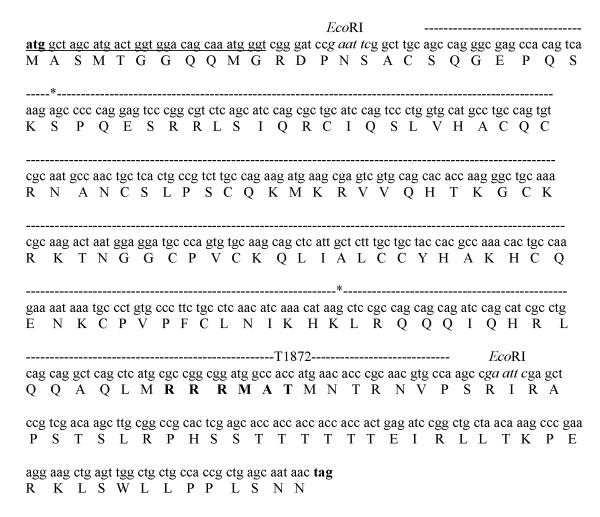
The Ala (A) or Asp (D) substitutions at CBP Thr-1872 were made by PCR mutagenesis, using the pCR®2.1 construct (Invitrogen; Carlsbad, CA) containing the CBP residues 1756-1878 (GenBank accession no. NM_001025432) as a DNA template. Although the source of this DNA is from a mouse species, the nucleotide sequence of the aforementioned CBP residues is 90% identical in mouse and human (GenBank accession no. NM_004380). The CBP fragment is flanked by EcoRI sites. Reverse primers for the T1872A and T1872D substitutions were 5′-GGCACGTTGCGGGTGTTCATGGCGGCCATCCGCCGGCGCATGAG-3′ and 5′-GGCACGTTGCGGGTGTTCATGTCGGCCATCCGCCGGCGCATGAG-3′, respectively. The bold nucleotides indicate the change in wildtype nucleotide sequence, while the underlined codons indicate the alanine and aspartic acid mutations. The forward primer used for the mutagenesis reaction was 5′-GCAGCCAGGGCGAGCCACAGTCAAAGAGCCCCCAGGAGTCACGC-3′. Following the amplification of the CBP fragment (373 nucleotides), EcoRI digestion was performed. The resultant DNA insert was ligated into a pET24b vector cut with EcoRI and dephosphorylated with calf intestinal alkaline phosphatase (New England Biolabs). Competent BL21(DE3) cells were transformed with individual ligation reactions. The correct insertion of substituted nucleotides and correct orientation of the mutant cDNAs were validated by DNA sequencing, using a T7 promoter primer of pET24b (Novagen) and an internal CBP primer (5′-AGCCACAGTCAAAGAGCCC-3′). During the construction of CBP CH3/pET24b, a single Trp at the C-terminus was incorporated to serve as an intrinsic probe of local dynamic changes. The confirmed sequence of the cloned CBP is shown in Fig. 2.
Fig. 2.
Nucleotide and protein sequences of cloned mouse CBP in pET24b. The start and stop codons within the multiple cloning sites, T7 tag, and EcoRI sites of pET24b are shown in bold, underlined, and italic nucleotides. The cloned CBP sequence, proposed Akt consensus site, and beginning and end of CH3 domain are indicated in dashed lines, bold residues, and asterisks, respectively.
Protein expression
The wildtype (WT) and mutant CBP CH3 fragments (T1872A or T1872D) were expressed in E. coli BL21(DE3)pLysS cells, which incorporate an 11-amino acid T7 tag sequence at the N-terminus. A 1L Luria Broth containing 30 μg/ml of kanamycin and 34 μg/ml of chloramphenicol were inoculated with 1 ml starter culture and grown at 37°C until OD600 ~0.5. The cell culture was then induced with a final concentration of 1 mM IPTG and grown for additional 12-16 hours. Following harvest by centrifugation at 4°C, the cell pellet was resuspended in BugBuster protein extraction reagent (Novagen) by pipetting and supplemented with protease cocktail inhibitor II (Calbiochem) and Benzonase® nuclease (Novagen), following manufacturer’s recommendations. Cell debris was removed by centrifugation at 12,000 rpm for 20 min, while the clarified supernatant was precipitated at 4°C with (NH4)2SO4 at 90% saturation. The resulting milky suspension was spun down at 12,000 rpm for 20 min at 4°C to yield a pellet, which was subsequently dissolved in 10 mM Bis-Tris Propane, 50 mM NaCl, 10 mM DTT, 1 mM EDTA, pH 7. Cell lysate was stored at −20°C until use or purified as described below. Total protein concentrations of cell lysates and fractionated peaks described below were determined via Bradford Protein Assay (Bio-Rad), using bovine serum albumin (BSA) as a protein standard.
Determination of isoelectric point of target protein
Based on previous futile attempts to purify the CH3 fragment of CBP by immunoprecipitation with monoclonal antibody against T7 Tag, we explored the possibility of isolating our expressed target protein by ionic charge. To determine which ion exchange media is appropriate, a 2-dimensional (2-D) gel electrophoresis was performed on cell lysate of WT CBP CH3 fragment using a ZOOM IPGRunner™ system (Invitrogen), following manufacturer’s recommendations. Parallel protein gels were run and subjected to either to Coomassie blue staining or Western blot analysis as described below. The experimental isoelectric point (pI) was manually determined, using the ruler tool of Kodak Imager software.
Western Blotting
Proteins resolved in a pre-cast Bis-Tris gel (Invitrogen) were electrotransferred to a PVDF membrane, followed by blocking in 10% BSA solution prepared in TBST (Tris-buffered saline with 0.1% Tween-20). This membrane was then probed separately with specific antibodies. In the case of proteins resolved by 2-D gel electrophoresis, the PVDF membrane was treated separately with HRP conjugate of T7 tag antibody at 1:5000 dilution and Akt substrate motif antibody at 1:1000 dilution. Following three 5-min washes in TBST, the separated proteins on a PVDF membrane were visualized by chemiluminescence using SuperSignal West Dura Extended Duration Substrate (Pierce) and a Kodak Image Station 2000MM.
Chromatographic separation of protein mixture
Purifications of the WT and mutant CBP CH3 fragments were performed in two chromatographic steps, using an AKTA FPLC (GE Healthcare). The first chromatography employed hydroxyapatite column (Bio-Rad) equilibrated in 10 mM Na2HPO4, pH 7, followed by linear gradient to 400 mM Na2HPO4, pH 7 at a flow rate of 1.0 ml/min. Fractionated peaks were analyzed by 4-12% gradient SDS-PAGE and SYPRO Ruby staining (Molecular Probes). Parallel Western blot analyses were performed with antibodies against T7 tag and Akt substrate motif. Fractions positively identified by Western blots near the theoretical molecular weight (MW) were pooled together and applied to a size exclusion column (Superdex 200) that had been equilibrated in 10 mM Bis-Tris Propane, 50 mM NaCl, 10 mM DTT, 1 mM EDTA, pH 7. Based on the elution profile of gel filtration standards and theoretical molecular weight of T7-tagged CBP fragment, the expected retention time was 30.7 min. Fractionated peaks obtained from the Superdex 200 were subjected to SDS-PAGE and Western blot analyses as described above. Each purification medium was cleaned up and regenerated as recommended by manufacturer.
Steady-state emission of WT CBP CH3 fragment
The spectral properties of WT CH3 were monitored by fluorescence spectroscopy in the absence or presence of GST-Akt1 fusion protein (Cell Signaling) at 1:1 and 10:1 molar ratios, using an excitation wavelength of 290 nm and slit widths of 10 nm via Shimadzu RF-1501 spectrofluorophotometer. Emission spectra were corrected for reference buffer, in this case 25 mM Tris, 5 mM DTT, pH 7.4 with 0.1% PEG 8000. The emission spectra of WT CBP CH3 with increasing GST-Akt1 concentrations were also determined.
Determination of equilibrium dissociation constants of WT and mutant CBP fragments
Equilibrium binding studies were performed to determine the relative affinities of CBP CH3 fragments for Akt1. Serial dilutions of equilibrated [Akt1•CBP CH3] complex, consisting initially of 40 nM Akt1 and 400 nM CBP CH3, were prepared in 25 mM Tris, 5 mM DTT, 0.1% PEG 8000, pH 7.4. Under such conditions, the observed spectral changes are primarily attributed to the emission of the CBP CH3 fragments. The steady-state emission of CBP CH3 was monitored at excitation wavelength of 290 nm at 25°C and corrected for the emission intensity of the reference buffer. Specifically, the interaction between CBP CH3 and Akt1 was monitored from the increase in intensity at 345 nm, which is within the wavelength range at which solvent-accessible Trp exhibits strong emission. The mole fraction of bound CBP CH3 was determined from the following ratio: x = (ICH3-Imax)/(ICH3-Icomplex), where ICH3, Imax, and Icomplex are the intensity for the unbound CBP CH3, observed maximum intensity, and intensity for the complex, respectively. The equilibrium dissociation constant (KD) for each CBP CH3 fragment was derived from the equation below via DeltaGraph 4 (Salt Lake City, UT), where v is the mole fraction of CBP CH3 bound to total protein and P and C are the upper and lower plateau, respectively:
Detection of protein-protein interaction via GST pull-down assay
The interaction between GST-Akt1 fusion protein and WT CBP CH3 fragment was confirmed by GST pull-down technique. Briefly, 500-μl reactions containing 5 μg of purified GST-Akt1 or S. japonicum GST (GenScript) were incubated with 50 μl of 50% slurry of Glutathione Sepharose beads (GE Healthcare) in 20 mM Tris/1 mM EDTA/100 mM NaCl, pH 7.4 (NET buffer) at 4°C for 1 hour with rotation. Beads were then centrifuged at 14,000 rpm for 2 min, followed by two washes with NET buffer. The immobilized GST-Akt1 or GST were incubated with equimolar amounts of WT CBP CH3 at 4°C for 2 hrs with rotation. Thereafter, beads were washed two times with NET buffer containing 0.5% Nonidet P-40. The pull-down complex was resuspended in reducing 1X sample buffer (Invitrogen), denatured by heating at 70°C for 10 min, and resolved by 8% SDS-PAGE. Pull-down assay was also performed with GSK-3α/β fusion protein (Cell Signaling), which is a known substrate of Akt1. This was followed by SYPRO Ruby (Invitrogen) staining to determine sizes and abundance of associated proteins. Parallel Western blots were also performed, using anti-T7 tag (Novagen) and anti-GST (GE Healthcare) antibodies to detect associated WT CBP CH3 and GSK-3α/β, respectively.
Phosphorylation of CBP CH3 fragment by Akt1
To determine whether the CH3 domain of CBP contains a phospho-acceptor site, we performed an in vitro kinase assay using the activated Akt1 fusion protein which contains an N-terminal GST tag (GST-Akt1). Briefly, 5 μg of wildtype or mutant CBP CH3 were reacted with 0.1 μg of GST-Akt1 in kinase buffer (25 mM Tris, 5 mM β-glycerophosphate, 2 mM DTT, 0.1 mM Na3VO4, 10 mM MgCl2, pH 7.5) containing 5 μCi of γ-32P-ATP (NEN) for 1 hr at 37°C. Parallel reactions without GST-Akt1 were also prepared. Reactions containing 1 μg of GSK-3α/β fusion protein (Cell Signaling) serve as controls. As additional control, 1 unit of protein phosphatase 2A (PP2A) was added to a reaction containing WT CBP CH3. Phosphorylated proteins were resolved by 4-12% SDS-PAGE and visualized by autoradiography.
Results
Experimental pI of recombinant CBP CH3 fragment is different from theoretical pI
Earlier attempts to purify the CBP CH3 were based on the assumption that the protein fragment has a theoretical pI of ~10.4, as determined by either Compute pI/MW or ProtParam tool (http://us.expasy.org/tools/), and thus be able to bind to a cation exchanger below its pI. However, none of the attempted cation exchangers were able to capture the target proteins at the recommended buffering conditions. To determine the actual pI of CBP CH3 in cellular lysates expressing the protein of interest, a 2-D protein gel electrophoresis was performed, followed by detection of the T7-tagged CBP CH3 protein via Western blot analysis. Several migrating proteins were observed as spots of varying intensity on the Coomassie blue-stained gel (Fig. 1B). Of these, only a few spots stained positive for the T7 tag (Fig. 1C) and Akt substrate motif (Fig. 1D). In both stained gel and parallel Western blots, only one spot was clearly positioned near the theoretical MW of the CBP CH3 fragment (MW 20.9 kDa), as indicated by a circle in Fig. 1B, 1C, and 1D. The experimental pI of this protein fragment (pI ~6.5) was approximately 3.9 units below its theoretical pI.
A two-step purification scheme was sufficient to yield a homogenous CBP CH3 fragment
We have employed two chromatographic steps to purify wildtype and mutant CBP CH3 fragments. All purification steps were performed at room temperature. The frozen bacterial lysate was thawed, spun down at 4°C to remove particulate matter, applied to a 5-ml sample loop (GE Healthcare) connected to a hydroxyapatite column, and then fractionated via an AKTA FPLC system. Hydroxyapatite has a chemical composition of Ca10(PO4)6(OH)2 [35] and binds to proteins via ionic interactions either between the positively charged calcium ions of the resin (Ca2+) and the negatively charged carboxylic side chains of polypeptide (COO−), or between the negatively charged phosphate ions (PO42−) of the resin and the positively charged amino side chains (NH3+) of polypeptide [36-38]. Given the experimental pI of the T7-tagged CBP CH3 fragment (Fig. 1B) and pH of the equilibration buffer, the protein fragment would have an overall net negative charge. As a result, the primary interaction of the protein fragment is through calcium ions. Bound proteins are then eluted by competitive binding with phosphate ions as indicated in Materials and methods.
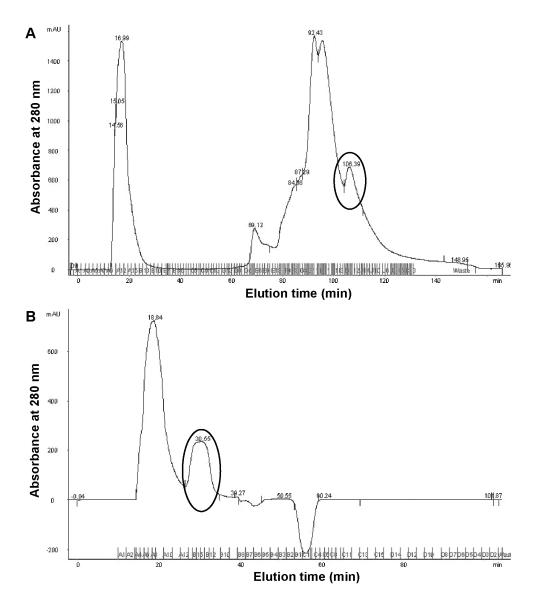
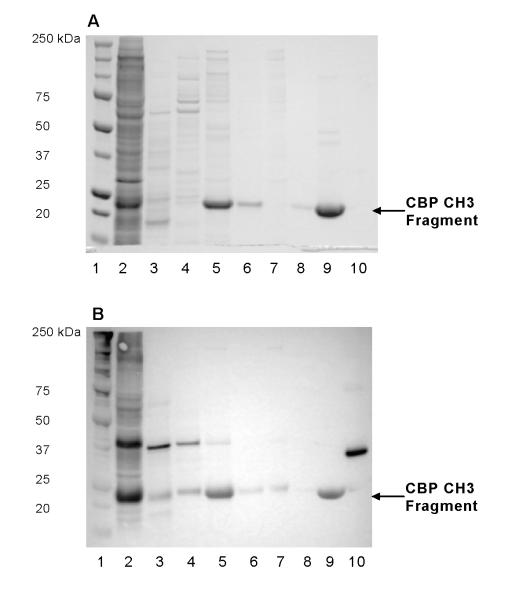
All fractions collected from hydroxyapatite column were monitored at absorbance of 280 nm (Fig. 3A) and characterized by SDS-PAGE and Western blotting (Fig. 4). Only fractions containing the T7 tag near the theoretical MW of CBP CH3 fragments were pooled together (Fig. 4, lane 5) for the size exclusion chromatography, as described in Materials and methods.
Fig. 3.
Elution profiles of clarified lysates of CBP CH3 via (A) hydroxyapatite chromatography and subsequent (B) size-exclusion chromatography of pooled fractions from previous purification step. Indicated in circles are peaks corresponding to T7-tagged CBP CH3 proteins as assessed by Western blotting (see Materials and methods and Fig. 4).
Fig. 4.
Progress of purification of CBP CH3 proteins as monitored by (A) Coomassie blue staining and (B) Western blotting against T7 tag. Arrows indicate the position of observed MW of T7-tagged CBP CH3 proteins. Lane 1: MW markers; lane 2: cell lysate; lane 3: Post HA A11-A13 fractions; lane 4: Post HA G7-G14 fractions; lane 5: Post HA H-I fractions; lane 6: FT of Post HA H-I fractions; lane 7: Post SEC A5-A10 fractions; lane 8: FT of Post SEC A13-B12 fractions; lane 9: Post SEC A13-B12 concentrate; lane 10: T7-tagged positive control (31.1 kDa). HA, hydroxyapatite; SEC, Superdex 200; FT, flow-through.
To determine the expected retention time of the CBP CH3 fragment in the Superdex 200 column, a calibration curve was derived from a plot of gel filtration standards of known molecular weight vs. retention volume (data not shown). As indicated in Fig. 3B, the protein fragment eluted at 30.55 min, which is near the calculated retention time of 30.7 min. Western blot analysis indicated that the pooled fractions contained T7-tagged protein (Fig. 4B, lane 9). The pooled fractions were concentrated via a Microcon® centrifugal filter device (MWCO 10,000 Da; Millipore), subjected to final dialysis in 10 mM Bis-Tris Propane, 50 mM NaCl, 10 mM DTT, 1 mM EDTA, pH 7 at 4°C, and stored at −20°C until use. Based on densitometric analysis of stained gels (Fig. 4A), the pooled CBP CH3 fragments are ~98% pure and have an observed MW of 22.6 kDa (data not shown). Typical protein recovery for purification of 1L culture or 35-ml cell lysate is about 6% (Table 1). We also confirmed the identity of the purified fragment by Western blotting against the C-terminus of CBP (C-20) at 1:250 dilution. A single band was found at the observed MW (data not shown).
Table 1.
Purification of WT CBP CH3 fragment from 1-L culture media.
| Fraction name | Fraction volume (ml) |
Total protein concentration (mg/ml) |
Total protein amount (mg) |
Amount of protein of interesta (mg) |
Percentage of recovery |
Fold enrichment |
|---|---|---|---|---|---|---|
| Cell lysate | 35 | 1.48 | 51.8 | 5.91 | 100 | — |
| Hydroxyapatite chromatography |
10.5 | 0.796 | 8.36 | 0.96 | 16 | — |
| Size-exclusion chromatography |
3 | 0.780 | 2.34 | 0.35 | 6 | 36.8 |
Calculated from densitometric analysis of Coomassie blue-stained gels and corresponded to T7-tagged WT CBP CH3.
Fluorescence emission of WT CBP CH3 fragment
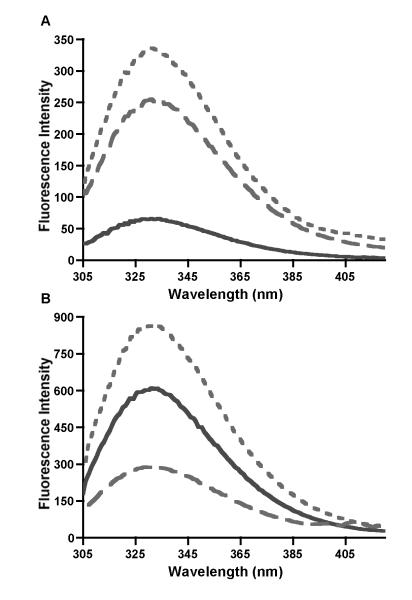
As indicated earlier, the recombinant fragment of WT CBP CH3 has one Trp incorporated at the C-terminus, which shows an average emission maximum at 330 nm (Fig. 5). Upon addition of increasing concentrations of Akt1 fusion protein (i.e., GST-Akt1) and incubation for 1 min at 25°C, the emission spectra showed a 10-fold increase in intensity but with no shift in wavelength maximum (data not shown). This increase in intensity is likely to be due to the intrinsic fluorescence of GST-Akt1, which has a single Trp in the GST tag sequence and 7 Trp in the Akt1 sequence. As shown in Fig. 5A (compare dashed and solid lines), the emission intensity of 10 nM GST-Akt1 is about 4.2-fold greater than that of 10 nM WT CBP CH3 alone, as expected for a protein with several Trp residues. The average wavelength maximum of GST-Akt1 (331 nm) is similar to that of WT CBP CH3. However, at 10:1 molar ratio of CBP CH3:GST-Akt1, the CBP fragment contributes ~73% of the total fluorescence (Fig. 5B, compare solid and dotted lines; compare dashed and dotted lines).
Fig. 5.
Representative emission spectra of wildtype CBP CH3 in the absence and presence of GST-Akt1. (A) Comparison of the emission spectra of 10 nM WT CBP CH3 (solid line), 10 nM GST-Akt1 (dashed line), and 1:1 molar ratio of WT CBP CH3 and GST-Akt1 (dotted line). (B) Emission spectra of 100 nM WT CBP CH3 (solid line), 10 nM GST-Akt1 (dashed line), and 10:1 molar ratio of the two proteins (dotted line).
Fluorescence titration of WT CBP CH3 with Akt1
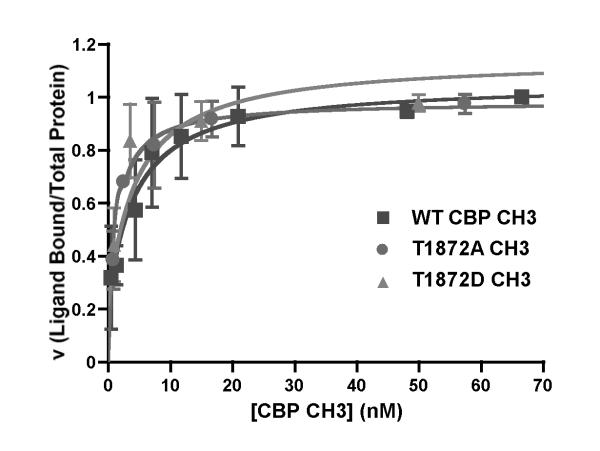
The intrinsic fluorescence signal of WT CBP CH3 was exploited to determine whether the association or dissociation between this protein fragment and GST-Akt1 can be monitored. At saturating concentrations of WT CBP CH3 or its Thr-1872 mutants, the fluorescence of the GST-Akt1•CBP CH3 complex is primarily due to Trp emission of the CBP CH3 fragment (Fig. 5B). Moreover, the titration curves for each protein were hyperbolic (Fig. 6), suggesting a single binding site of each protein fragment on Akt1 [39]. The equilibrium dissociation constants (KD) of WT CBP CH3, T1872A CH3, and T1872D CH3 for Akt1 are 4.30 nM, 1.20 nM, and 4.07 nM, respectively. These fairly similar KD values indicate that Ala or Asp mutations at Thr-1872 do not significantly alter the binding affinities of each CH3 fragment for Akt1.
Fig. 6.
Equilibrium binding titrations of wildtype and mutant CBP CH3 with Akt1. Titration curves for WT CBP CH3 (squares), T1872A CH3 (circles), and T1872D CH3 (triangles) were derived from equation described in Materials and methods. Dissociation constants (KD) of WT CBP CH3, T1872A CH3, and T1872D CH3 for activated Akt1 are 4.30 ± 0.10 nM, 1.20 ± 0.01 nM, and 4.07 ± 0.67 nM, respectively.
In vitro interaction between WT CBP CH3 and Akt1
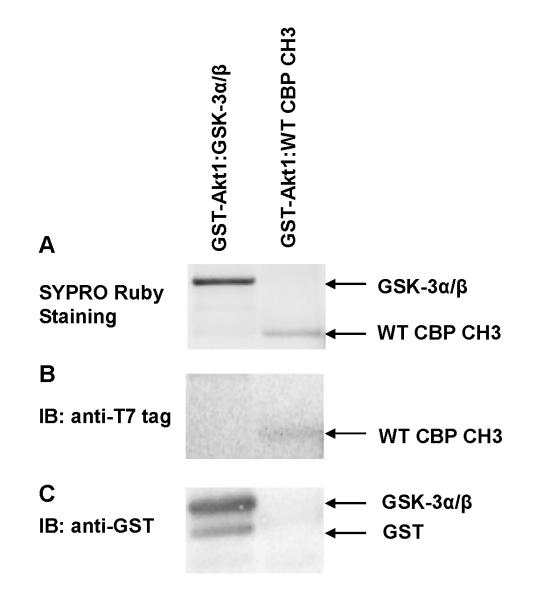
To determine whether or not WT CBP CH3 fragment binds directly to Akt1 and confirm our fluorescence binding studies, GST pull-down assays were performed. As indicated in Fig. 7A, WT CBP CH3 (MWobs ~22.6 kDa) was able to form a complex with GST-Akt1 compared to control GST. On the basis of densitometric scanning of stained gels (Fig. 7A and Table 2), the level of pull-down complex with WT CBP CH3, however, is 27-fold lower than with GSK-3α/β (MWobs ~27 kDa). This result shows that WT CBP CH3 interacts directly with Akt1, albeit its interaction is weaker than that of GSK-3α/β. To confirm that the associated protein with GST-Akt1 is CBP CH3 fragment, parallel Western blot analyses were performed with antibodies against T7 tag. Figure 7B shows a visible band corresponding to T7-tagged CBP CH3 fragment, indicating successful capture by GST-Akt1. The identity of GSK-3α/β fusion protein was also confirmed by Western blotting (Fig. 7C), including its N-terminal GST tag (MWobs ~26 kDa).
Fig. 7.
In vitro interactions between WT CBP CH3 or GSK-3α/β fusion protein and GST-Akt1 as monitored by (A) SYPRO Ruby staining and (B and C) Western blotting. Control reactions containing GST were also performed to confirm the specificity of WT CBP CH3:Akt1 interaction (data not shown). In this case, no WT CBP CH3 was pulled down from the control reactions (data not shown). Immunoblotting with anti-T7 tag antibody (B) was specific only for WT CBP CH3 fragment, while GST antibody immunoblotting (C) was specific for both GSK-3α/β and GST.
Table 2.
Protein recovery from GST pull-down assays
| Eluted Proteins from GSH Sepharose | Percentage of recoveryb |
|---|---|
| WT CBP CH3 | 1.67 ± 0.67 |
| GSK-3α/β | 31.7 ± 13.5 |
| Akt1 | 34.4 ± 2.0 |
Densitometric scanning of observed bands in stained gels relative to band density of starting amount of each protein used in the assays.
CBP fragment spanning the CH3 domain contains a phospho-acceptor site(s)
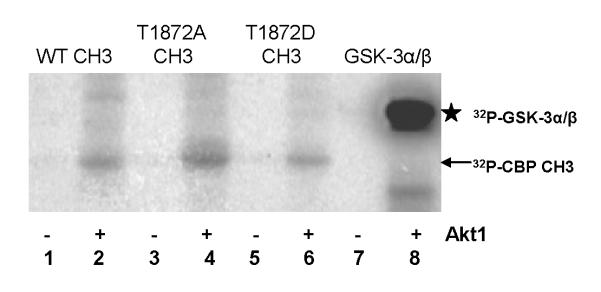
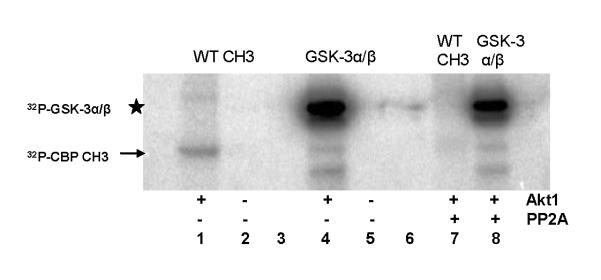
While the aforementioned in vitro interaction studies confirm GST-Akt1•CBP CH3 interaction, these studies do not determine the phosphorylation status of the suspected phospho-acceptor site in the CBP CH3 fragment, Thr-1872. Here, we determined whether Thr-1872 serves as the sole Akt1-mediated phosphorylation site, as suggested by others based on Akt consensus motif [24, 26, 27, 34]. As indicated in Fig. 8 (lanes 2, 4, and 6), radioactive 32P was incorporated in both WT and mutant CBP CH3 fragments in the presence of Akt1, suggesting that Thr-1872 is not the exclusive site of phosphorylation. This result is consistent with our equilibrium binding studies, which indicate indiscriminate binding of CBP CH3 fragments for Akt1. In contrast, the phosphorylation of GSK-3α/β was 3-fold greater than any of the CBP CH3 fragments (Fig. 8, lane 8). To confirm that the observed phosphorylated band of the WT CBP CH3 is not a residual effect of the assay conditions, parallel incubations with protein phosphatase 2A (PP2A) were performed. One unit of PP2A is sufficient to abrogate the phosphorylation of WT CBP CH3 fragment (Fig. 9, compare lanes 1 and 7). In comparison, this amount of PP2A resulted in 1.8-fold decrease in phosphorylation of GSK-3α/β (Fig. 9, compare lanes 4 and 8), while no change in a reaction containing either Akt1 alone or Akt1 with PP2A, as expected for unphosphorylatable T308D/S473D Akt1 (data not shown).
Fig. 8.
In vitro phosphorylation assays of CBP CH3 fragments and GSK-3α/β fusion protein in the absence and presence of Akt1. 32P-labeled proteins were identified in PVDF membrane by autoradiography. Arrow indicates the location of 32P-CBP CH3 fragments, while star indicates the location of 32P-GSK-3α/β.
Fig. 9.
Akt-mediated phosphorylation of WT CBP CH3 fragment is abolished by PP2A. Assay was performed as in Fig. 8 but in the presence of 1 unit of PP2A.
Discussion
Molecular interactions play a central role in the proper functioning of all biological systems [40-42]. Having an understanding of the structural and functional importance of protein-protein, protein-peptide, and protein-ligand interactions represents an opportunity for drug discovery or biomarker development. Fluorescence spectroscopy is a powerful tool for monitoring and elucidating interactions between biomolecules [43-45], given its high sensitivity in dilute solutions and availability of the equipment.
In the present study, we have exploited the intrinsic fluorescent properties of Trp in a recombinant fragment of an important nuclear coactivator of steroid/thyroid signaling, CBP, to study its potential protein-protein interaction with Akt1 in vitro. To our knowledge, this is the first in vitro study to demonstrate the association between CBP and cell survival factor Akt by fluorescence spectroscopy and phosphorylation of a protein fragment spanning the CH3 domain and part of the Q-rich domain of CBP by Akt1.
To study the potential molecular interaction between the C-terminal fragment of CBP and Akt1 in greater detail, we have undertaken a large-scale expression of the CBP fragment containing a T7 tag at the N-terminus and Trp at the C-terminus. Here, we report for the first time the successful purification of a non-GST fusion tag of CBP by ion-exchange chromatography that resulted in relatively homogenous proteins (Fig. 4). In particular, CBP CH3 fragment was primarily captured via Ca2+•••COO− coordination between the positively charged calcium ions of the hydroxyapatite resin and the negatively charged carboxylic groups of the acidic protein fragment [35-37]. The resulting purified CBP CH3 fragment has an observed MW (22.6 kDa) that is nearly 2 kDa larger than expected. This discrepancy in MW may be attributed to yet to be identified secondary structure at either the N-terminal T7 tag or the last 43 C-terminal residues (Fig. 2) and will be addressed in future structural studies. Nonetheless, the purified CBP CH3 fragment will serve as a valuable tool for future studies to better understand the role of CBP interaction with other proteins in cancer progression.
Full activation of Akt1 requires phosphorylation of Ser-473 and Thr-308 at the activation loop of the kinase domain and hydrophobic motif of the regulatory domain, respectively [46, 47]. To mimic this activation in our in vitro assays, a commercially available GST fusion protein containing the full-length Akt1 with T308D/S473D mutations was used. This Akt double mutant was found to be constitutively active and sufficient to phosphorylate a number of substrate targets [48, 49]. The binding of CBP CH3 fragment to activated Akt1 resulted in a significant increase in Trp fluorescence, perhaps due to a decreased solvent exposure of the fluorophore(s) in either CBP CH3 or Akt1 at the site of contact (Fig. 5 and 6). While our data demonstrates interaction between CBP CH3 fragment and Akt1 (Fig. 5, 6, and 7), the specific sites of contact between these two proteins cannot be deduced from our studies. Detailed structural analysis of this complex by either mass spectrometry or NMR is warranted.
While Ser-1834 of human p300 has been shown to be an Akt phosphorylation site [26, 27], it is not known whether the corresponding residue in CBP is the exclusive site of phosphorylation (Fig. 1A). Our data shows that the CBP CH3 fragment is phosphorylated by Akt in vitro (Fig. 8 and Fig. 9), and this phosphorylation event is reversed by PP2A (Fig. 9). However, Thr-1872 is not the exclusive site of phosphorylation, as the removal of phosphorylatable Thr (T1872A) or substitution with unphosphorylatable Asp (T1872D) at residue 1872 of CBP CH3 fragment does not block phosphorylation by activated Akt1. This result suggests an alternative phosphorylation site(s) in the CBP fragment.
The discrepancy between our studies and previous studies [26, 27] is presumably due to differences in the surface accessibility (Table 3) of the Akt consensus motifs in human p300 (1.031) and CBP (0.789), as determined by Motif Scan (http://scansite.mit.edu/index.html), resulting in a slightly higher probability of Akt-mediated phosphorylation of p300 S1834 (0.193%) compared to CBP T1871 (0.171%). Upon closer inspection of the sequence of human CBP C-terminus (GenBank accession no. NM_004380), Ser-1771 shows a higher probability of Akt1-mediated phosphorylation (0.396%) compared to Thr-1871 (0.171%) at medium stringency (Table 3). Similar result was obtained with the mouse CBP (GenBank accession no. NM_001025432), considering that CBP is highly homologous in human and mouse (data not shown).
Table 3.
Predicted phosphorylation sites in the C-terminus of human p300 and CBP
| Protein | Akt consensus sequence |
Predicted site | Probability of phosphorylation by Akt relative to other protein kinases |
Surface accessibility |
|---|---|---|---|---|
| p300 | RRRMAS | S1834 | 0.193% | 1.031 |
| CBP | RRRMATc | T1871 | 0.171% | 0.789 |
| CBP | ESRRLSc | S1771 | 0.396% | 1.179 |
Corresponding residues in mouse CBP are T1872 and S1772.
Based on the surface accessibility of the predicted Akt-mediated phosphorylation sites (Table 3), human CBP Thr-1871 (or mouse CBP T1872) appears to be buried compared to Ser-1771 (or mouse CBP S1772), and thus has a lower probability of coming in contact with Akt1. Whether this biochemical reaction occurs is not known, given the important limitation of the calculation of surface accessibility. In this case, the structural feature of the entire CBP C-terminus, which is yet to be determined, is not taken into account. Perhaps, detailed examination of the rapid kinetics of Akt1•CBP CH3 interaction and subsequent phosphorylation of CBP CH3 fragment will provide better understanding of the catalytic mechanism of Akt1. Future studies will look into the in vitro as well as in vivo phosphorylation status of CBP Ser-1772 and its site-directed mutants.
In summary, our studies provide the first report of successful purification of CBP fragment with naturally fluorescent Trp for binding studies and the first direct evidence for in vitro phosphorylation of CBP C-terminal fragment by Akt1. Given the roles of both Akt and CBP in human cancers and the availability of agents targeting the PI3K/Akt/mTOR signaling, we aim to delineate the exact contribution of in vivo Akt1-mediated phosphorylation of CBP to breast cancer progression and development of drug resistance in the future. Finally, our high-throughput fluorescence-based approach to study protein-protein interaction may open new avenues for drug or biomarker development.
Acknowledgments
We thank Dr. Julie Ostrander and Dr. Victoria Seewaldt for helpful discussion, and Robin Marjoram and Dr. Kellye Kirkbride for technical assistance. This work was supported in part by grants from the National Institute of Health/National Cancer Institute: 2P30CA14236 (Career Development to C.I.D.) and Minority Research Supplement to R01CA088799 (V.L. Seewaldt).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
- CREB
- 3′,5′-cyclic adenosine monophosphate response element binding protein
- CBP
- CREB-binding Protein
- HAT
- histone acetyl transferase
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- CaMKIV
- Ca2+/calmodulin-dependent protein kinase IV
- IKKα
- IκB kinase alpha
- AP-1
- activating protein-1
- NF-κB
- nuclear factor-kappaB
- MMP-9
- matrix metalloproteinase-9
- PKB
- Protein Kinase B
- PI3K
- phosphatidylinositol 3-kinase
- mTOR
- mammalian target of rapamycin
- HMECs
- human mammary epithelial cells
- Tam
- tamoxifen
- Bis-Tris propane
- 1,3-Bis[tris(hydroxymethyl)methylamino]propane
- DTT
- DL-dithiothreitol
- EDTA
- ethylenediamine-tetraacetic acid
- IPTG
- isopropyl-β-D-thiogalactopyranoside
- GST
- glutathione S-transferase
- HRP
- horseradish peroxidase
- BSA
- bovine serum albumin
- FPLC
- fast phase liquid chromatography
- SDS-PAGE
- sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- PP2A
- protein phosphatase 2A
- NMR
- nuclear magnetic resonance.
References
- [1].Chakravarti D, LaMorte VJ, Nelson MC, Nakajima T, Schulman IG, Juguilon H, Montminy M, Evans RM. Role of CBP/p300 in nuclear receptor signaling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- [2].Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- [3].Chan HM, La Thangue NB. p300/CBP proteins: HAT for transcriptional bridges and scaffolds. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- [4].Goodman RH, Smolik SM. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- [5].Kung AL, Rebel VI, Bronson RT, Ch’ng LE, Sieff CA, Livingston DM, Yao TP. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- [6].Kalkhoven E. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- [7].Timmermann S, Lehrmann H, Polesskaya A, Harel-Bellan A. Histone acetylation and disease. Cell Mol. Life Sci. 2001;58:728–736. doi: 10.1007/PL00000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Horvai AE, Xu L, Korzus E, Brard G, Kalafus D, Mullen TM, Rose DW, Rosenfeld MG, Glass CK. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc. Natl. Acad. Sci. USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wadgaonkar R, Phelps KM, Haque Z, Williams AJ, Silverman ES, Collins T. CREB-binding protein is a nuclear integrator of nuclear factor-kappaB and p53 signaling. J. Biol. Chem. 1999;274:1879–1882. doi: 10.1074/jbc.274.4.1879. [DOI] [PubMed] [Google Scholar]
- [10].Adamson AL, Kenney S. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with histone acetylase CREB-binding protein. J. Virol. 1999;73:6551–6558. doi: 10.1128/jvi.73.8.6551-6558.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ghosh AK, Yuan W, Mori Y, Chen Sj., Varga J. Antagonistic regulation of type I collagen gene expression by interferon-gamma and transforming growth factor-beta. Integration at the level of p300/CBP transcriptional coactivators. J. Biol. Chem. 2001;276:11041–11048. doi: 10.1074/jbc.M004709200. [DOI] [PubMed] [Google Scholar]
- [12].Dietze EC, Caldwell LE, Marcom K, Collins SJ, Yee L, Swisshelm K, Hobbs KB, Bean GR, Seewaldt VL. Retinoids and retinoic acid receptors regulate growth arrest and apoptosis in human mammary epithelial cells and modulate expression of CBP/p300. Microsc. Res. Tech. 2002;59:23–40. doi: 10.1002/jemt.10174. [DOI] [PubMed] [Google Scholar]
- [13].Tsai LN, Ku TK, Salib NK, Crowe DL. Extracellular signals regulate rapid coactivator recruitment at AP-1 sites by altered phosphorylation of both CREB binding protein and c-jun. Mol. Cell. Biol. 2008;28:4240–4250. doi: 10.1128/MCB.01489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jordan CV, O’Malley BW. Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. J. Clin. Oncol. 2007;25:5815–5824. doi: 10.1200/JCO.2007.11.3886. [DOI] [PubMed] [Google Scholar]
- [15].Gamble MJ, Freedman LP. A coactivator code for transcription. Trends in Biochem. Sci. 2002;27:165–167. doi: 10.1016/s0968-0004(02)02076-5. [DOI] [PubMed] [Google Scholar]
- [16].Lonard DM, O’Malley BW. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–414. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- [17].Ait-Si-Ali S, Carlisi D, Ramirez S, Upegui-Gonzalez LC, Duquet A, Robin P, Rudkin B, Harel-Bellan A, Trouche D. Phosphorylation by p44 MAP Kinase/ERK1 stimulates CBP histone acetyl transferase activity in vitro. Biochem. Biophys. Res. Commun. 1999;262:157–162. doi: 10.1006/bbrc.1999.1132. [DOI] [PubMed] [Google Scholar]
- [18].Impey S, Fong AL, Wang Y, Cardinaux JR, Fass DM, Obrietan K, Wayman GA, Storm DR, Soderling TR, Goodman RH. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34:235–244. doi: 10.1016/s0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- [19].Zanger K, Radovick S, Wondisford FE. CREB binding protein recruitment to the transcription complex requires growth factor-dependent phosphorylation of its GF box. Mol. Cell. 2001;7:551–558. doi: 10.1016/s1097-2765(01)00202-7. [DOI] [PubMed] [Google Scholar]
- [20].Huang WC, Ju TK, Hung MC, Chen CC. Phosphorylation of CBP by IKKα promotes cell growth by switching the binding preference of CBP from p53 to NF-κB. Mol. Cell. 2007;26:75–87. doi: 10.1016/j.molcel.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Janknecht R, Nordheim A. MAP kinase-dependent transcriptional coactivation by Elk-1 and its cofactor CBP. Biochem. Biophys. Res. Commun. 1996;228:831–837. doi: 10.1006/bbrc.1996.1740. [DOI] [PubMed] [Google Scholar]
- [22].Hardingham GE, Chawla S, Cruzalegui FH, Bading H. Control of recruitment and transcription-activating function of CBP determines gene regulation by NMDA receptors and L-type calcium ions. Neuron. 1999;22:789–798. doi: 10.1016/s0896-6273(00)80737-0. [DOI] [PubMed] [Google Scholar]
- [23].Gusterson R, Brar B, Faulkes D, Giordano A, Chrivia J, Latchman D. The transcriptional co-activators CBP and p300 are activated via phenylephrine through the p42/p44 MAPK cascade. J. Biol. Chem. 2002;277:2517–2524. doi: 10.1074/jbc.M104626200. (2002) [DOI] [PubMed] [Google Scholar]
- [24].Kovacs KA, Steinmann M, Magistretti PJ, Halfon O, Cardinaux JR. CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J. Biol. Chem. 2003;278:36959–36965. doi: 10.1074/jbc.M303147200. [DOI] [PubMed] [Google Scholar]
- [25].Gusterson RJ, Yuan LW, Latchman DS. Distinct serine residues in CBP and p300 are necessary for their activation by phenylephrine. Int. J. Biochem. Cell Biol. 2004;36:893–899. doi: 10.1016/j.biocel.2003.10.004. [DOI] [PubMed] [Google Scholar]
- [26].Guo S, Cichy SB, He X, Yang Q, Ragland M, Ghosh AK, Johnson PF, Unterman TG. Insulin suppresses transactivation by CAAT/enhancer-binding proteins beta (C/EBPbeta). Signaling to p300/CREB-binding protein by protein kinase B disrupts interaction with the major activation domain of C/EBPbeta. J. Biol. Chem. 2001;276:8516–8523. doi: 10.1074/jbc.M008542200. [DOI] [PubMed] [Google Scholar]
- [27].Huang WC, Chen CC. Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyl transferase and transcriptional activity. Mol. Cell. Biol. 2005;25:6592–6602. doi: 10.1128/MCB.25.15.6592-6602.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kandel ES, Hay N. The regulation and activities of the multifunctional serine/threonine kinase Akt/PKB. Exp. Cell Res. 1999;253:210–229. doi: 10.1006/excr.1999.4690. [DOI] [PubMed] [Google Scholar]
- [29].Bellacosa A, Kumar CC, Di Cristofano A, Testa JR. Activation of AKT kinases in cancer: implications for therapeutic targeting. Adv. Cancer Res. 2005;94:29–86. doi: 10.1016/S0065-230X(05)94002-5. [DOI] [PubMed] [Google Scholar]
- [30].De Guzman RN, Liu HY, Martinez-Yamout M, Dyson HJ, Wright PE. Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP. J. Mol. Biol. 2000;303:243–253. doi: 10.1006/jmbi.2000.4141. [DOI] [PubMed] [Google Scholar]
- [31].Dietze EC, Caldwell LE, Grupin SL, Mancini M, Seewaldt VL. Tamoxifen but not 4-hydroxytamoxifen initiates apoptosis in p53(-) normal human mammary epithelial cells by inducing mitochondrial depolarization. J. Biol. Chem. 2001;276:5384–5394. doi: 10.1074/jbc.M007915200. [DOI] [PubMed] [Google Scholar]
- [32].Dietze EC, Troch MM, Bean GR, Heffner JB, J.B., Bowie ML, Rosenberg P, Ratliff B, Seewaldt VL. Tamoxifen and tamoxifen ethyl bromide induce apoptosis in acutely damaged mammary epithelial cells through modulation of AKT activity. Oncogene. 2004;23:3851–3862. doi: 10.1038/sj.onc.1207480. [DOI] [PubMed] [Google Scholar]
- [33].Dietze EC, Bowie ML, Mrozek K, Caldwell LE, Neal C, Marjoram RJ, Troch MM, Bean GR, Yokoyama KK, Ibarra CA, Seewaldt VL. CREB-binding protein regulates apoptosis and growth of HMECs grown in reconstituted ECM via laminin-5. J. Cell Sci. 2005;118:5005–5022. doi: 10.1242/jcs.02616. [DOI] [PubMed] [Google Scholar]
- [34].Sanchez M, Sauve K, Picard N, Tremblay A. The hormonal response of estrogen receptor beta is decreased by the phosphatidylinositol 3-kinase/Akt pathway via a phosphorylation-dependent release of CREB-binding protein. J. Biol. Chem. 2007;282:4830–4840. doi: 10.1074/jbc.M607908200. [DOI] [PubMed] [Google Scholar]
- [35].Kawasaki T, Takahashi S, Ikeda K. Hydroxyapatite high-performance liquid chromatography: column performance for proteins. Eur. J. Biochem. 1985;152:361–371. doi: 10.1111/j.1432-1033.1985.tb09206.x. [DOI] [PubMed] [Google Scholar]
- [36].Gorbunoff MJ. The interaction of proteins with hydroxyapatite, I. Role of protein charge and structure. Anal. Biochem. 1984;136:425–432. doi: 10.1016/0003-2697(84)90239-2. [DOI] [PubMed] [Google Scholar]
- [37].Gorbunoff MJ, Timasheff S. The interaction of proteins with hydroxyapatite, III. Mechanism. Anal. Biochem. 1984;136:440–445. doi: 10.1016/0003-2697(84)90241-0. [DOI] [PubMed] [Google Scholar]
- [38].Kawasaki T. Hydroxyapatite as a liquid chromatographic packing. J. Chromatogr. 1991;544:147–184. [Google Scholar]
- [39].Eftink MR. Fluorescence methods for studying equilibrium macromolecule-ligand interactions. Methods Enzymol. 1997;278:221–257. doi: 10.1016/s0076-6879(97)78013-3. [DOI] [PubMed] [Google Scholar]
- [40].Jones S, Thornton JM. Principles of protein-protein interactions. Proc. Natl. Acad. Sci. USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- [42].Cusick ME, Klitgord N, Vidal M, Hill DE. Interactome: gateway into systems biology. Hum. Mol. Genet. 2005;14:R171–R181. doi: 10.1093/hmg/ddi335. [DOI] [PubMed] [Google Scholar]
- [43].Lakowicz JR. Protein Fluorescence. In: Lakowicz JR, editor. Principles of Fluorescence Spectroscopy. Kluwer Academic/Plenum Publishers; New York: 1999. pp. 445–486. [Google Scholar]
- [44].Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- [45].Viallet PM, Vo-Dinh T. Monitoring intracellular proteins using fluorescence techniques: from protein synthesis and localization to activity. Curr. Protein Pept. Sci. 2003;4:375–388. doi: 10.2174/1389203033487045. [DOI] [PubMed] [Google Scholar]
- [46].Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- [47].Ananthanarayanan B, Fosbrink M, Rahdar M, Zhang J. Live-cell molecular analysis of Akt activation reveals roles for activation loop phosphorylation. J. Biol. Chem. 2007;282:36634–36641. doi: 10.1074/jbc.M706227200. [DOI] [PubMed] [Google Scholar]
- [48].Bellacosa A, Chan TO, Ahmed NN, Datta K, Malstrom S, Stokoe D, McCormick F, Feng J, Tsichlis P. Akt activation by growth factors is a multi-step process: the role of the PH domain. Oncogene. 1998;17:313–325. doi: 10.1038/sj.onc.1201947. [DOI] [PubMed] [Google Scholar]
- [49].Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]