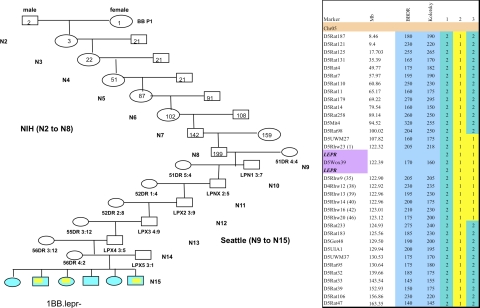
Fig. 1.
Breeding strategy (left) and genotyping of chromosome 5 (right). Left: the breeding approach of total N15 backcrosses that led to the generation of the new congenic line. The first intercross and from N2 to N8 backcrosses were generated in the laboratory of Dr. J. T. Hansen at the National Institutes of Health (NIH). From N9 to N15 backcrosses were produced in the Robert H. Williams Laboratory, University of Washington, of Å. Lernmark. At each backcross the DNA was sent to Seattle for genotyping assessment. Numbers and names in and below each box represent rat ID. Right: simple sequence length polymorphism (SSLP) marker map of chromosome 5. Marker annotations are shown together with the University of California-Santa Cruz (UCSC) map marker location and the size of the marker in the 3 parental strain BioBreeding diabetes-resistant (BBDR) and Koletsky rats. Purple, localization of the Lepr gene expanded from 122.320.075 bp to 122.503.437 bp with the marker D5Wox39 inside the DNA fragment that comprised the gene; blue, markers polymorphic between the parental strains. Animals in lanes 1–3 represent the parental and new Lepr−/− congenic line. The animal in lane 1 is diabetes resistant (DR), lane 2 Koletsky (Kol), lane 3 BBDR.Lepr− (D5Rat98-D5Rat233)Rhw. The numerical color code is green (2) DR/DR and yellow (1) Kol/Kol. Colors in left panel correspond with colors in lanes 1 and 3 in right panel.