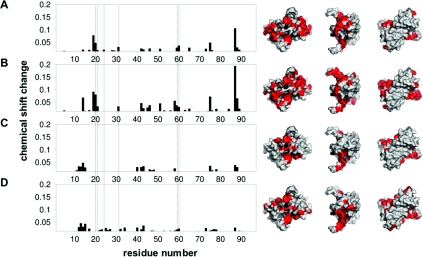
Figure 2. NMR chemical-shift analysis of the Alba1 interaction with nucleic acids.
The weighted sum of 15N-r and 1H- proton chemical shift changes (√[(Δδn/10)2+Δδnh2]) upon titration of Alba with (A) 16 bp dsDNA, (B) 16 mer ssDNA, (C) 16 mer ssRNA and (D) 28A ssRNA are plotted against residue number. Vertical broken lines show residues associated with a conserved crystallographic dimer–dimer interface. Those residues with a sum chemical-shift change >0.015 are plotted. These are highlighted in red on the corresponding surface representations of Alba1 in order to indicate the regions that are perturbed upon binding. Lys16 and Arg44, which are in intermediate exchange, are shown in dash grey. Lys16 has been shown previously to be involved in binding DNA [7], and Arg44 is likely to be involved given its position and charge. The structural representations were generated using MacPyMOL (DeLano Scientific; http://www.pymol.org).