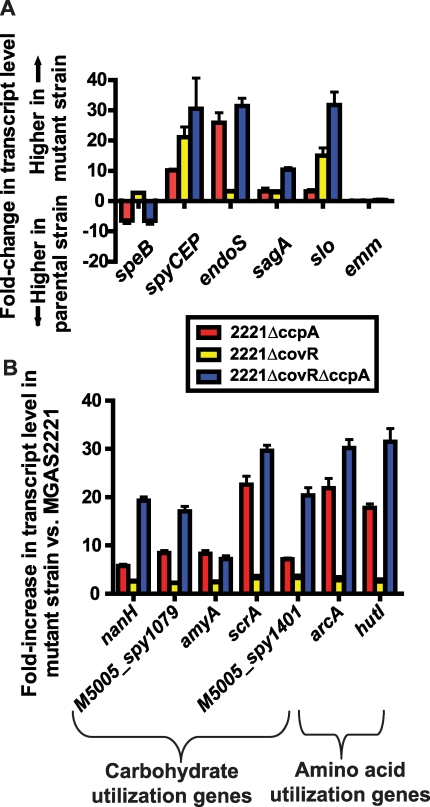
Figure 4. Identification of significant overlap between the CcpA and CovR transcriptomes.
Differences in gene transcript levels measured by expression microarray analysis for select GAS virulence factor encoding genes (A) and genes involved in carbohydrate and amino acid utilization (B). Data graphed are mean +/− standard deviation for quadruplicate samples for strains as indicated in the insert box. All transcript levels are significantly different compared to strain MGAS2221 except for the emm gene which is shown for reference purposes. A significant difference was defined as at least two-fold difference in median gene transcript level and P<0.05 for the indicated isogenic mutant strain compared to the parental wild-type strain MGAS2221. speB, streptococcal pyrogenic exotoxin B; spyCEP, Streptococcus pyogenes cell envelope proteinase; endoS, endoglycosidase S; sagA, streptolysin S; slo, streptolysin O; emm, M protein; nanH, acetylneuraminate lyase; amyA, cyclomaltodextrin glucanotransferase; scrA, sucrose transport enzyme; arcA, arginine deiminase; hutI, imidazolonepropionase.