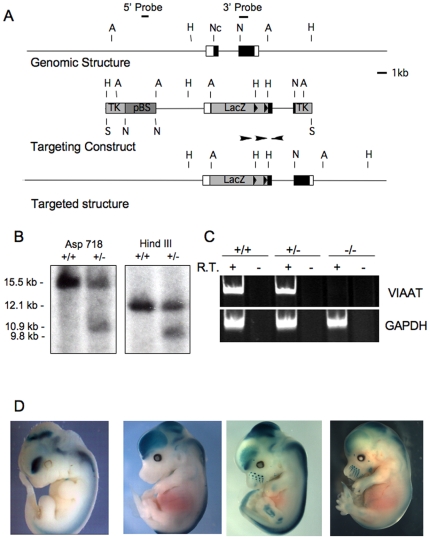
Figure 1. Targeted disruption of the Viaat gene in mice.
(A) A schematic representation of the Viaat wild-type genomic locus (Genomic Structure), the targeting vector (Targeting Construct), and the mutant locus (Targeted structure) are shown. A LacZ sequence was inserted into an NcoI site at the Viaat start codon. The lox sites that flank the Neo resistance cassette are indicated with two arrowheads in the region immediately 3′ to the lacZ sequence. The locations of the 5′ (5′ Probe) and 3′ (3′ Probe) flanking probes used in characterizing the Viaat allele are indicated above the map of the wild type Viaat locus. The positions of the restriction sites used in the Southern blot analysis of genomic DNA from ES cells are indicated by single letters. Restriction sites are indicated as follows: H (HindIII), Nc (NcoI), A (Asp718) and N (NotI). (B) Southern blot analysis of genomic DNA from a wild type parental ES cell line (+/+) and a targeted ES cell line used to generate the ViaatlacZ mouse (+/-) are shown. In genomic DNA digested with Asp718 the 3′flanking probe hybridized to a 15.5 kb wild type fragment and a 10.9 kb from the targeted allele (Asp718 panel) and in genomic DNA digested with HindIII this probe hybridized to a 12.1 kb wild type band and a 9.8 kb mutant band (HindIII panel). (C) RT-PCR analysis of cDNA from wild-type (+/+) , heterozygous (+/−), and mutant embryo brain at E16.5. For each genotype RT-PCR was performed on RNA that had been reverse transcribed (+ lanes) and RNA that had not been incubated with reverse transcriptase (−) to control for non-specific amplification. GAPDH primers were used as a positive control. (D) Localized expression of β-galactosidase activity in ViaatlacZ and Gad1lacZ heterozygotes. The panels from left to right show β-galactosidase activity in E11.5, and E14.5 ViaatlacZ embryos and in E12.5 and E14.5 Gad1lacZ embryos.