Abstract
A neuroscientific experiment typically generates a large amount of data, of which only a small fraction is analyzed in detail and presented in a publication. However, selection among noisy measurements can render circular an otherwise appropriate analysis and invalidate results. Here we argue that systems neuroscience needs to adjust some widespread practices in order to avoid the circularity that can arise from selection. In particular, “double dipping” – the use of the same data set for selection and selective analysis – will give distorted descriptive statistics and invalid statistical inference whenever the results statistics are not inherently independent of the selection criteria under the null hypothesis. To demonstrate the problem, we apply widely used analyses to noise data known not to contain the experimental effects in question. Spurious effects can appear in the context of both univariate activation analysis and multivariate pattern-information analysis. We suggest a policy for avoiding circularity.
Introduction
“Show me the data,” we say. But we don’t mean it. Instead of the numbers generated by measurement – which can be billions for a single experiment – we wish to see results. This frequent confusion illustrates an important point: We think of the results as reflecting the data – so closely that we can disregard the distinction. However, interposed between data and results is analysis; and analysis is often complex and always based on assumptions (Fig. 1a, top).
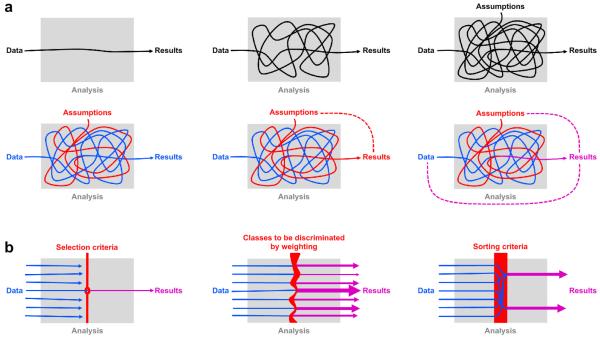
Fig. 1. Intuitive diagrams for understanding circular analysis.
(a) The top row serves to remind us that our results reflect our data indirectly: through the lens of an often complicated analysis, whose assumptions are not always fully explicit. The bottom row illustrates how the assumptions (and hypotheses) can interact with the data to shape the results. Ideally (bottom left), the results reflect some aspect of the data (blue) without distortion (although the assumptions will determine what aspect of the data is reflected in the results). But sometimes (bottom center) a close inspection of the analysis reveals that the data get lost in the process and the assumptions (red) predetermine the results. In that case the analysis is completely circular (red dotted line). More frequently in practice (bottom right), the assumptions tinge the results (magenta). The results are then distorted by circularity, but still reflect the data to some degree (magenta dotted lines). (b) Three diagrams illustrate the three most common causes of circularity: selection (left), weighting (center), and sorting (right). Selection, weighting, and sorting criteria reflect assumptions and hypotheses (red). Each of the three can tinge the results, distorting the estimates presented and invalidating statistical tests, if the results statistics are not independent of the criteria for selection, weighting, or sorting.
Ideally, the results reflect some aspect of the data without any distortion caused by assumptions or hypotheses (Fig. 1a, bottom left). Consider the hypothesis that neuronal responses in a particular region reflect the difference between two experimental stimuli. We might measure the neuronal responses, average across repetitions, and present the results in a bar graph with one bar for the response to each stimulus. The set of stimuli (or, more generally, experimental conditions) is decided on the basis of assumptions and hypotheses, thus determining what bars are shown. But the results themselves, i.e. the heights of the two bars, are supposed to reflect the data without any effect of assumptions or hypotheses.
Untangling how data and assumptions influence neuroscientific analyses sometimes reveals that assumptions predetermine results to some extent.1,2,3,4,5 When the data are altogether lost in the process, the analysis is completely circular (Fig. 1a, bottom center). More frequently, in practice, the results do reflect the data, but are distorted – to varying degrees – by the assumptions (Fig. 1a, bottom right). Such distortions can arise when the data are first analyzed to select a subset, and then the subset is reanalyzed to obtain the results. In this context, assumptions and hypotheses determine the selection criterion, and selection, in turn, can distort the results.
In neuroimaging, an example of selection is the definition of a region of interest (ROI) by means of a statistical mapping that highlights voxels more strongly active during one condition than another. In single-cell recording, an example of selection is the restriction of in-depth analysis to neurons with certain response properties. In electro- and magnetoencephalography, an example of selection is the restriction to a subset of sensors or sources that show expected responses.
In gene microarray studies, an example of selection is inferential analysis performed for a statistically selected subset of genes.6
In behavioral studies, an example of selection is the division of a group of subjects into subgroups based on task performance. Weighting and sorting of data can be construed as variants of selection; and we will use the latter term in a general sense to refer to all three (Fig. 1b).
Selection can entail two distinct forms of bias: (1) selective reporting of accurate results and (2) distortion of estimates and invalidation of statistical tests. Both forms deserve a wider debate, but this paper focuses on the latter.
If selection were determined only by true effects in the data, there would be no distortion of the results of the selective analysis. However, data are always a composite of true effects and noise. Selection, thus, is affected by noise. In neuroimaging, for example, the voxels included at the fringe of an ROI tend to reflect the noise to some extent – even if the ROI highlights a truly active brain region (as in Example 2, below). When the selection process is based on the design matrix, it creates spurious dependencies between the noise in the selected data and the experimental design, thus violating the assumption of random sampling. This can bias selective analysis.
Selective analysis is a powerful tool and perfectly justified whenever the results are statistically independent of the selection criterion under the null hypothesis. However, “double dipping” – the use of the same data for selection and selective analysis – will result in distorted descriptive statistics and invalid statistical inference whenever the test statistics are not inherently independent of the selection criteria under the null hypothesis. Nonindependent selective analysis is incorrect and should not be acceptable in neuroscientific publications.
Although the dangers of double dipping in the pool of data are well understood in statistics and computer science, the practice is common in systems neuroscience, and in particular in neuroimaging and electrophysiology. To assess how widespread nonindependent selective analyses are in the literature, we examined all functional-magnetic-resonance-imaging (fMRI) studies published in five prestigious journals (Nature, Science, Nature Neuroscience, Neuron, Journal of Neuroscience) in 2008. Of these 134 fMRI papers, 42% (57 papers) contained at least one nonindependent selective analysis (not considering supplementary materials). Another 14% (20 papers) may contain nonindependent selective analyses, but the methodological information given was insufficient to reach a judgment.
Are all these studies incorrect in their main claims? We do not think so. First, we counted any study containing at least one nonindependent selective analysis. For a given paper, the overall claim may not depend on the distorted result. Second, we have no way of assessing the severity of the distortions. They might be small in many cases.
If circularity consistently caused only slight distortions, one could argue that it is a statistical quibble. However, the distortions can be very large (Example 1, below) or smaller, but significant (Example 2); and they can affect the qualitative results of significance tests. In order to decide which neuroscientific claims hold, the community needs to carefully consider each particular case – guided by neuroscientific as well as statistical expertise. Reanalyses and replications may also be required.
The problem arises so frequently, because the desired selection criterion is often identical with or – however subtly – related to the desired results statistics for the selective analysis. In neuroimaging, for example, we may hypothesize that there is a region responding more strongly to stimulus A than B, select voxels showing this effect to define an ROI, and then selectively analyze that ROI to test our hypothesis. One way to ensure statistical independence of the results under the null hypothesis is to use an independent data set for the final analysis of the selected channels (e.g. neurons or voxels).
Another way to ensure independence is to use inherently independent statistics for selection and selective analysis. For example, we may select channels with a large average response to stimuli A and B (contrast A+B) and test for a difference between the conditions (contrast A-B). The contrast vectors ([1 1]T and [1 -1]T) are orthogonal. Unfortunately, contrast-vector orthogonality, by itself, is not sufficient to ensure independence (see Supplementary Information: A policy for noncircular analysis, Fig. S3). In practice, the same data are frequently used for selection and selective analysis, even when the selection criteria are not inherently independent of the results statistics. In that case, the results are questionable.
Distortions arising from selection tend to make results look more consistent with the selection criteria, which often reflect the hypothesis being tested. Circularity therefore is the error that beautifies results – rendering them more attractive to authors, reviewers, and editors, and thus more competitive for publication. These implicit incentives may create a preference for circular practices, as long as the community condones them.
Analyzing multiple channels and reporting results for a statistically selected subset is essential in electrophysiology and neuroimaging. Neuroimaging is faced with even more parallel sites than electrophysiology – typically on the order of 100,000 voxels within the measured volume. However, selection is also an issue in electrophysiology and will gain importance as multi-electrode arrays become more widely used. To its great credit, neuroimaging has developed rigorous methods for statistical mapping from the beginning.7,8,9,10,11 Note that mapping the whole measurement volume avoids selection altogether: We can analyze and report results for all locations equally, while accounting for the multiple tests performed across locations.12 The sense of discovery associated with brain mapping derives from this data-driven approach, which avoids both the bias of selective reporting of accurate results and the circularity that can invalidate nonindependent selective analyses. Despite the beauty and completeness of a nonselective mapping analysis, selective in-depth analysis of ROIs can yield additional insights.13
In this paper, we demonstrate the problem using two examples from neuroimaging (Figs. 2, 3). In each example, a widely accepted practice is applied to random data known not to contain the experimental effect in question. This exercise reveals the distortion and spurious significance that can arise in circular analysis. We view the problem from three perspectives: as ‘selection bias’, as ‘exploration and confirmation using the same data’, and as ‘overfitting’. These perspectives are elaborated on in the Supplementary Information, which also contains further analyses and simulations (Figures S1-S4), and a comprehensive set of questions and answers about circular analysis. Finally, we suggest a policy for noncircular analysis of brain-activity data (Fig. 4, Supplementary Discussion).
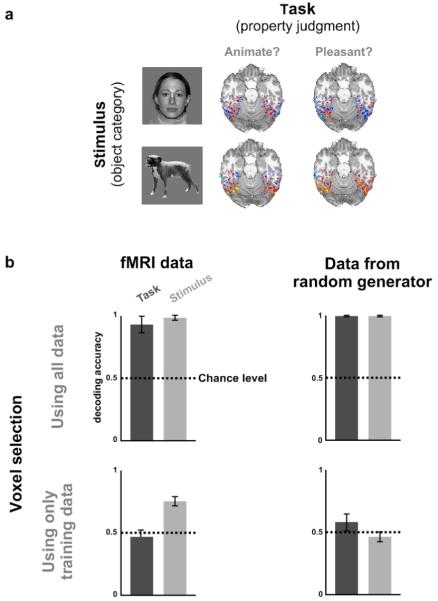
Fig. 2. Example 1: Data selection can bias pattern-information analysis.
(a) In order to assess to what extent human inferior-temporal activity patterns reflect bottom-up sensory signals and top-down task constraints, we measured activity patterns with fMRI while subjects viewed object images of different categories and judged either whether the object shown was “animate” (task 1) or whether it was “pleasant” (task 2).2 (b) We selected all inferior-temporal voxels for which any two-sided t test contrasting two conditions was significant at p<0.001 (uncorrected for multiple tests). We then cleanly divided the data by using odd runs for training and even runs for testing. We used a linear classifier to determine whether the activity pattern would allow us to decode the stimulus category (light gray bars) and the judgment task (dark gray bars). Results (top left) suggested that both stimulus and task can be decoded with high accuracy, significantly above chance. However, application of the same analysis to Gaussian random data (top right), also suggested high decoding accuracies significantly above chance. This shows that spurious effects can appear when data from the test set is used in the initial data-selection process. Such spurious effects can be avoided by performing selection using data independent of the test data (bottom row). Error bars indicate +/−1 across-subject standard error of the mean. For details on experiment and analysis, see Example 1: Pattern-information analysis.
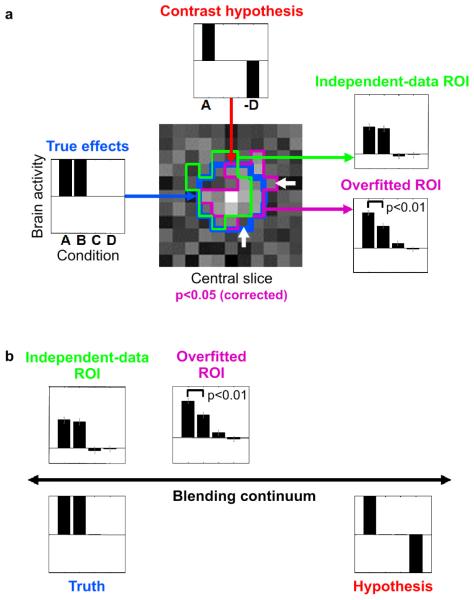
Fig. 3. Example 2: ROI definition can bias activation analysis.
A simulated fMRI block-design experiment demonstrates that nonindependent ROI definition can distort effects and produce spuriously significant results, even when the ROI is defined by rigorous mapping procedures (accounting for multiple tests) and highlights a truly activated region. Error bars indicate +/− 1 standard error of the mean. (a) The layout of this panel matches the intuitive diagrams of Fig. 1a: The data in Fig. 1a correspond to the true effects (left); the assumptions to the contrast hypothesis (top), and the results to ROI-average activation analyses (right). A 100-voxel region (blue contour in central slice map) was simulated to be active during conditions A and B, but not during conditions C and D (left). The t map for contrast A-D is shown for the central slice through the region (center). When thresholded at p<0.05 (corrected for multiple tests by a cluster threshold criterion), a cluster appears (magenta contour), which highlights the true activated region (blue contour). The ROI is somewhat affected by the noise in the data (difference between blue and magenta contours). The noise pushes some truly activated voxels below the threshold and lifts some nonactivated voxels above the threshold (white arrows). This can be interpreted as overfitting. The bar graph for the overfitted ROI (bottom right, same data as used for mapping) reflects the activation of the region during conditions A and B as well as the absence of activation during conditions C and D. However, in comparison to the true effects (left) it is substantially distorted by the selection contrast A-D (top). In particular, the contrast A-B (simulated to be zero) exhibits spurious significance (p<0.01). When we use independent data to define the ROI (green contour), no such distortion is observed (top right). For details on the simulation and analysis, see Example 2: Regional activation analysis in the text. (b) The simulation illustrates how data selection blends truth (left) and hypothesis (right) by distorting results (top) so as to better conform to the selection criterion.
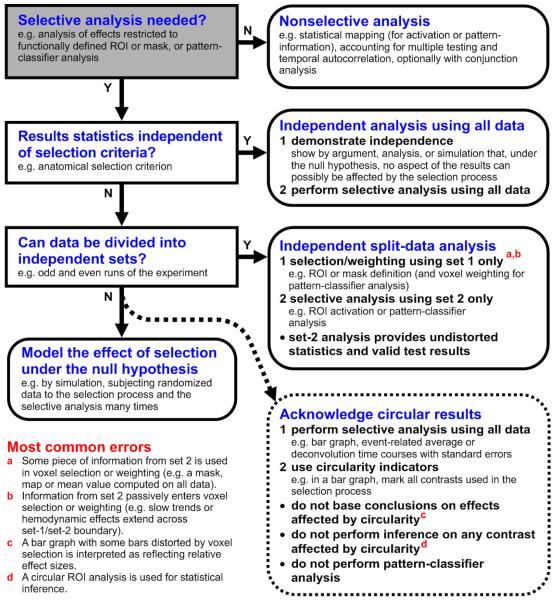
Fig. 4. A policy for noncircular analysis.
This flow diagram suggests a procedure for choosing an appropriate analysis that avoids the pitfalls of circularity. Considering the most common errors (bottom left, red letter references) can help recognize circularity in assessing a given analysis. The gist of the policy is as follows: We first consider performing a nonselective analysis only. If selective analysis is needed and we can demonstrate that the results are independent of the selection criterion under the null hypothesis, then all data are used for selective analysis. If we cannot demonstrate this, then a split-data analysis can serve to ensure independence. (For details, see Supplementary Information, A policy for noncircular analysis.)
Example 1: Pattern-information analysis
In pattern-information analysis,14,15,16,17,18 the objective is to determine whether the pattern of response in a brain region contains stimulus information. Considering pattern-information analysis is relevant not only because this approach is gaining importance in systems neuroscience, but also because it provides a powerful general perspective on circular analysis.19,20
One popular approach to pattern-information analysis is to attempt to decode the stimulus from the response pattern with a pattern classifier.21,22,23 If we can “predict” the stimuli from the response patterns significantly above chance level, then the patterns must contain information about the stimuli. The most common method is linear classification, where a linear decision boundary (i.e. a hyperplane) is placed in response-pattern space to discriminate the stimuli. After training the classifier to discriminate example patterns, we can determine its accuracy (percentage of correct classifications). However, if we used the training data to assess the accuracy, we would overestimate the accuracy and conclude that there is stimulus information even if there is none. The reason for this is a phenomenon known as “overfitting”: A model will capture the noise to some extent as its parameters are fitted to the data. A more flexible model (i.e. one with many parameters) will tend to be more susceptible to overfitting. However, even the fitting of a one-parameter model (e.g. a mean) is affected by noise to some extent. When thinking about fitting a linear decision boundary, we tend to imagine a line separating two clouds of points in a plane. When there are many points (much data) and few dimensions (e.g. two dimensions: a plane), overfitting may be negligible. However, response-pattern space has as many dimensions as there are response channels (e.g. neurons or voxels); and a linear decision boundary has as many parameters as there are dimensions. Counter to the intuitive simplicity and rigidity of a planar decision boundary, fitting a hyperplane in a 100 dimensional space in order to separate 100 data points is like separating two points on a plane by a line: separation is always perfect – even if the points are drawn from identical distributions (Supplementary Information: Overfitting of model parameters). Separability, thus, provides no evidence for separate distributions.
Using the same data to train and test a linear classifier can lead us to believe that there is information about the stimulus in regions where actually there is none. In this context, double dipping entails extreme distortions and is widely understood to be unacceptable. We are not aware of examples of this error in the systems neuroscience literature. However, the error here is fundamentally the same as that of nonindependent selective analysis. Linear classification is based on a weighted sum of the responses. Weighting can be construed as a continuous variant of selection. Conversely, we can think of selection as binary weighting, a special case.
Can selection produce similar distortions as continuous weighting in the context of pattern-information analysis? In order to test this possibility, we performed a classifier analysis on human inferior-temporal response patterns measured with fMRI while subjects viewed object images.2 The experiment had two independent variables: object category and task (Fig. 2a). In task 1, subjects judged whether the object presented was animate or inanimate. In task 2, they judged whether the object was pleasant or unpleasant. The experiment can reveal to what extent inferior-temporal activity patterns reflect stimulus category and task.
We first analyzed all experimental runs together to define an ROI. We included all inferior-temporal voxels for which any two-sided t test for a pairwise condition contrast was significant at p<0.001 (uncorrected). We then cleanly divided the data into independent training and test sets by designating all odd runs as training data and all even runs as test data. For training and test set separately, we computed the average activity pattern for each condition (combination of task and stimulus category). For each pair of conditions, we decoded a given test pattern by assigning the condition label of the training pattern more similar to the test pattern.14 This nearest-neighbor method is a linear classifier, because the condition-average patterns are used. Pattern similarity was measured by the Pearson correlation across voxels. For each subject, decoding accuracy was computed (a) for each pairwise task comparison within each stimulus category and (b) for each pairwise stimulus-category comparison within each task (chance level: 50%). Task decoding accuracies were averaged, first within subjects and then across subjects. Stimulus-category decoding accuracies were averaged in the same way. Similar methods are widespread in the literature.
This analysis suggested that both stimulus category and judgment task can be decoded with accuracies above 90% and significantly better than chance (Fig. 2b, top left). So we would conclude that the task as well as the stimulus category is strongly reflected in inferior-temporal response patterns.
However, when we applied the same analysis to data generated with a Gaussian random generator, we obtained equivalent results (Fig. 2b, top right). The random data are known not to contain any information about either task or stimulus category, so any correct analysis should indicate decoding accuracies whose deviations from 50% are within the margin of error and come up significant in only 5% of the cases. This demonstrates that selection of ROI voxels using all data can strongly bias estimates of decoding accuracy and yield spuriously significant test results.
The cause of the distortion is the selection of voxels whose time series, by chance, exhibit some consistency between training and test set in the way they are related to the experimental conditions. For the selected voxel set, thus, training and test data sets are no longer independent.
When we corrected the error of nonindependent voxel selection, decoding accuracies dropped to chance level for the Gaussian random data (Fig. 2b, bottom right). For the actual experimental data, task decoding accuracy dropped to chance level, whereas stimulus-category decoding accuracy dropped to about 75% but remained significant (Fig. 2b, bottom left). The latter result replicates a previous study.14
Beyond neuroimaging, pattern-information analyses are increasingly used in invasive and scalp electrophysiology. Circularity will cause similar distortions when cells or sensors are preselected by nonindependent criteria.
We conclude that selection of response channels can strongly inflate estimates of decoding accuracy and misleadingly suggest substantial amounts of information in a brain region, where actually there is none. We can avoid such spurious results by performing selection using data independent of the test data.
Example 2: Regional activation analysis
A widespread approach to neuroimaging analysis is to perform a statistical mapping, followed by a selective activation analysis of one or more ROIs. The ROIs are typically defined by the mapping; and their analysis is often based on the same data. In many cases, the conclusion that the ROI analysis serves to support is directly or indirectly related to the mapping contrast. Is this a valid approach?
Let us assume that the ROI is defined by a valid statistical mapping analysis with adequate correction for multiple tests. (If the statistical mapping were not performed correctly, one could argue that whatever problem arises thereafter is not caused by nonindependent selection, but by the inadequate statistical mapping.) We further assume that the mapping analysis successfully localizes a truly active region. (The alternative case that the mapping falsely highlights a region, will be rare – it will have a probability of 0.05 or less under the null hypothesis, since the mapping is assumed to be correct. If the mapping did not highlight any region, then there would be no ROI to selectively analyze.)
In order to assess whether an ROI analysis can be distorted by selection under these assumptions, we simulated a neuroimaging data set of 30 by 30 by 20 voxels and 200 time points. The simulated experiment was a block design with four conditions (A, B, C, D). We placed a 100-voxel activation (5 by 5 by 4 voxels) at the center of the volume. The region was simulated to be active during conditions A and B, but not C and D (Fig. 3a, left). The resulting spatiotemporal data set was added to independent spatiotemporal Gaussian noise and spatially smoothed by convolution with a 3-voxel-wide cubic kernel. The data were analyzed by means of a general linear model using the same design matrix as used to simulate the effects, with one predictor per condition. We mapped the data set by voxelwise univariate linear modeling using the contrast A-D (Fig. 3a, top). We thresholded the resulting t map using a primary threshold corresponding to p<0.0001, uncorrected. We then assessed the size of each contiguous cluster exceeding this primary threshold and highlighted all clusters whose size exceeded a cluster-size threshold that controlled the familywise error rate at p<0.05, thus correcting for multiple tests. (The cluster-size threshold was determined by simulating the map-maximum-cluster-size distribution under the null hypothesis by running the above simulation 1000 times for the same contrast without any effect placed in the data.)
The ROI defined by the mapping analysis (Fig. 3a, magenta contour) correctly highlights the activated region (blue contour). However, the ROI is somewhat affected by noise in the data (difference between blue and magenta contours). Some voxels at the fringe of the ROI (white arrows) will be included because their noise component makes them look as though they conformed slightly better to the selection criterion; others will be excluded because their noise makes them look as though they did not conform as well to the selection criterion (magenta contour and map in the background). This can be interpreted as overfitting of the ROI.
We now average all time courses within the ROI (same data as used for mapping) and fit the linear model. The resulting bar graph (Fig. 3a, bottom right) reflects the activation of the region during conditions A and B as well as the absence of activation during conditions C and D. However, it is substantially distorted by the nonindependent selection: Recall that the mapping was based on the contrast A-D (Fig. 3a, top). Although the region is equally activated during conditions A and B, it appears to be more activated during condition A than B; and this effect is significant (p<0.01 in the particular example run shown). When we use independent data to define the ROI (green contour), no such distortion is observed (Fig. 3a, top right).
In order to assess the proportion of cases, in which the contrast A-B would yield a spuriously significant result caused by non independent voxel selection, we repeated the simulation 100 times. The one-sided t test for the ROI contrast A-B (whose ground-truth value is zero in the simulation) was significant in 20 of the 100 simulations for p<0.05 and in 9 of the 100 simulations for p<0.01. These false-positives rates are significantly larger than for a correct test (p=0.00005, χ2 test for the null hypothesis that the proportion of p<.05-significant results is 0.05). We conclude that nonindependent selection can distort the results of selective analyses, even when rigorous statistical tests are used during selection.
Independence of the selective analysis could have been ensured either by using independent test data (Fig. 3a, top right) or by using selection and test statistics that are inherently independent. For the contrasts used (selection contrast: A-D, test contrast: A-B), the inherent dependence is obvious: Voxels with higher signals during condition A are more likely to be selected by chance using contrast A-D; thus test contrast A-B will be biased. However, selection bias can arise even for orthogonal contrast vectors (Supplementary Information: A policy for noncircular analysis, Fig. S3).
Nonindependent selection causes bias, because the selection is somewhat affected by the noise (difference between blue and magenta ROIs, Fig. 3a), even when the statistical criterion is stringent and the ROI highlights a truly activated region. Our statistical selection method controls the familywise error rate; it does not ensure that the ROI perfectly captures the shape of the region. The ROI will be overfitted to the data to some extent – just like the weights of a linear classifier.
To temper this conclusion, we note that overfitting will typically be less severe in fitting an ROI than in fitting a linear classifier with continuous weights: The restriction to binary weights and the constraint of selecting a contiguous set of voxels effectively regularize an ROI fit. By contrast, discontiguous selection (as in Example 1, above) and data sorting can be extremely susceptible to overfitting. (For two simple simulations on sorting effects, see Fig. S2.)
In practice, statistical mapping for ROI definition is not always performed with rigorous correction for multiple tests as assumed here. Many studies rely on a threshold of p<.001, uncorrected. The selective analysis of the same data is then sometimes interpreted as though it confirmed the effect selected for. While it does not confirm the effect, the selective analysis effectively serves to help us forget about the multiple-testing problem during selection. The inadequacy of the inference during selection will compound the circularity of the selective analysis and strong biases as well as large false-positives rates are to be expected.
Although the example here concerns the selection of voxels in a neuroimaging experiment, the same caution should be applied in analyzing other types of data. In single-cell recording, for example, it is common to select neurons according to some criterion (e.g. visual responsiveness or selectivity) before applying further analyses to the selected subset. If the selection is based on the same data set as used for selective analysis, biases will arise for any statistic not inherently independent of the selection criterion. For neurons as well as voxels, selection should be based on criteria independent of any selective analysis.
In sum, Example 2 shows that nonindependent selective analysis can cause significant biases, even when selection is performed with rigorous statistical inference correcting for multiple tests.
A policy for noncircular analysis
One possible policy that ensures correct inference and undistorted descriptive statistics is summarized by the flow diagram of Fig. 4. The core of our policy is as follows: we first consider a nonselective analysis (e.g. brain mapping with correction for multiple comparisons). If selective analysis is needed, we next assess whether the results statistics are independent of the selection criterion under the null hypothesis. If this has been explicitly demonstrated, then all data are used for selective analysis. Otherwise, an independent data set is used for the selective analysis to ensure independence of the results under the null hypothesis and prevent circularity. Each of these steps is explained in detail in the Supplementary Information under A policy for noncircular analysis.
Conclusion
In order to learn about brain function, systems neuroscience needs to apply complex selective and recurrent analyses to high-dimensional brain-activity data. One challenge this poses is to avoid circularity. A circular analysis is one whose assumptions distort its results. We have demonstrated that practices widespread in neuroimaging are affected by circularity. In particular, data weighting, sorting, and selection can distort results and invalidate tests when preceding nonindependent further analyses. Similar practices are common in other fields of systems neuroscience including electrophysiology. The distortions may be small in many cases. However, they can be large and can qualitatively affect results. We conclude that some common practices need to be adjusted. In particular, selection criteria should be demonstrated to be independent of further analyses. A simple way to ensure independence is to use independent data for selection and selective analyses. Immanuel Kant24 observed that Reason, in science, will not be led on by Nature, but rather forces her to answer specific questions. Circular analysis goes one step further, enforcing specific answers as well (or biasing results in their favor) – one step too far in our opinion.
Supplementary Material
Acknowledgments
We would like to thank P. A. Bandettini, R. W. Cox, J. V. Haxby, D. J. Kravitz, A. Martin, R. A. Poldrack, R. D. Raizada, Z. S. Saad, J. T. Serences, and E. Vul for helpful discussions. This work was supported by the Intramural Research Program of the US National Institute of Mental Health.
References
- 1.Baker CI, Hutchison TL, Kanwisher N. Does the fusiform face area contain subregions highly selective for nonfaces? Nat Neurosci. 2007;10(1):3–4. doi: 10.1038/nn0107-3. [DOI] [PubMed] [Google Scholar]
- 2.Simmons WK, Matlis S, Bellgowan PS, Bodurka J, Barsalou LW, Martin A. Imaging the context-sensitivity of ventral temporal category representations using high-resolution fMRI. Society for Neuroscience Abstracts. 2006 [Google Scholar]
- 3.Baker CI, Simmons WK, Bellgowan PS, Kriegeskorte N. Circular inference in neuroscience: The dangers of double dipping. Society for Neuroscience Abstracts. 2007 doi: 10.1038/nn.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vul E, Kanwisher N. Begging the question: The non-independence error in fMRI data analysis. In: Hanson S, Bunzl M, editors. Foundations and Philosophy for Neuroimaging. in press. To appear in. [Google Scholar]
- 5.Vul E, Harris C, Winkielman P, Pashler H. Perspectives on Psychological Science. Puzzlingly High Correlations in fMRI Studies of Emotion, Personality, and Social Cognition. in press. [DOI] [PubMed] [Google Scholar]
- 6.Benjamini Y. Comment: Microarrays, Empirical Bayes and the Two-Groups Model. Statistical Science. 2008;23(1):23–28. [Google Scholar]
- 7.Worsley KJ, Evans AC, Marrett S, Neelin P. A three-dimensional statistical analysis for CBF activation studies in human brain. J. Cereb. Blood Flow Metab. 1992;12:900–918. doi: 10.1038/jcbfm.1992.127. [DOI] [PubMed] [Google Scholar]
- 8.Friston KJ, Jezzard P, Turner R. Analysis of functional MRI time-series. Hum. Brain Mapp. 1994;1:153–171. [Google Scholar]
- 9.Worsley KJ, Friston KJ. Analysis of fMRI time-series revisited – again. Neuroimage. 1995;2:173–181. doi: 10.1006/nimg.1995.1023. [DOI] [PubMed] [Google Scholar]
- 10.Nichols TE, Hayasaka S. Controlling the Familywise Error Rate in Functional Neuroimaging: A Comparative Review. Statistical Methods in Medical Research. 2003;12(5):419–446. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- 11.Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. NeuroImage. 2002;15(4):870–8. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- 12.Friston KJ, Rotshtein P, Geng JJ, Sterzer P, Henson RN. A critique of functional localisers. NeuroImage. 2006;30(4):1077–87. doi: 10.1016/j.neuroimage.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 13.Saxe R, Brett M, Kanwisher N. Divide and conquer: a defense of functional localizers. NeuroImage. 2006;30(4):1088–96. doi: 10.1016/j.neuroimage.2005.12.062. [DOI] [PubMed] [Google Scholar]
- 14.Haxby JV, Gobbini MI, Furey ML, Ishai A, Schouten JL, Pietrini P. Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science. 2001;293:2425–2430. doi: 10.1126/science.1063736. [DOI] [PubMed] [Google Scholar]
- 15.Cox DD, Savoy RL. Functional magnetic resonance imaging (fMRI) “brain reading”: detecting and classifying distributed patterns of fMRI activity in human visual cortex. Neuroimage. 2003;19:261–270. doi: 10.1016/s1053-8119(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell TM, Hutchinson R, Niculescu RS, Pereira F, Wang X. Learning to decode cognitive states from brain images. Machine Learning. 2004;57:145–175. [Google Scholar]
- 17.Kriegeskorte N, Goebel R, Bandettini P. Information-based functional brain mapping. Proc. Natl. Acad. Sci. USA. 2006;103:3863–3868. doi: 10.1073/pnas.0600244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norman KA, Polyn SM, Detre GJ, Haxby JV. Beyond mind-reading: multi-voxel pattern analysis of fMRI data. Trends Cogn. Sci. 2006;10:424–430. doi: 10.1016/j.tics.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Strother SC, Anderson J, Hansen LK, Kjems U, Kustra R, Sidtis J, Frutiger S, Muley S, LaConte S, Rottenberg D. The Quantitative Evaluation of Functional Neuroimaging Experiments: The NPAIRS Data Analysis Framework. Neuroimage. 2002;15:747–771. doi: 10.1006/nimg.2001.1034. [DOI] [PubMed] [Google Scholar]
- 20.Strother S, La Conte S, Hansen LK, Anderson J, Zhang J, Pulapura S, Rottenberg D. Optimizing the fMRI data-processing pipeline using prediction and reproducibility performance metrics: I. A preliminary group analysis. Neuroimage. 2004;23:S196–207. doi: 10.1016/j.neuroimage.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell TM. Machine Learning. McGraw Hill; 1997. [Google Scholar]
- 22.Duda RO, Hart PE, Stork DG. Pattern Classification. 2nd Edition Wiley; 2001. [Google Scholar]
- 23.Bishop CM. Pattern Recognition and Machine Learning. Springer; 2006. [Google Scholar]
- 24.Kant I. Kritik der reinen Vernunft. 2nd Edition. 1787. http://www.gutenberg.org/dirs/etext04/8ikc210.txt. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.