Fig. 4.
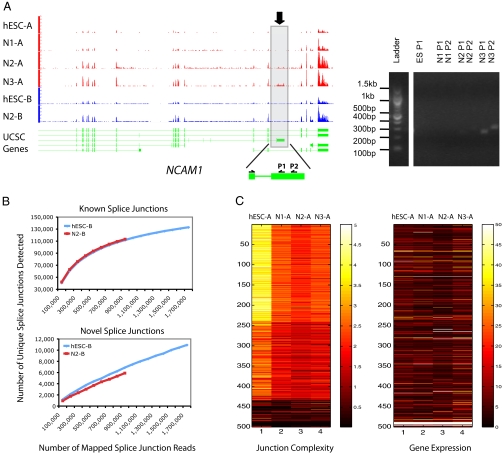
Splicing analysis. (A) One of the transcript isoforms of neural cell adhesion molecule 1 (NCAM1) (marked by a rectangle and arrow) is primarily expressed in N3 and very weakly at N2 but not at N1 and hESC stages. The y axis of the RNA-Seq signal tracks represents the read density normalized by the number of mapped reads per million for each cell type. Two sets of RT-PCR primers were designed on the alternative exon and the adjacent constant exon, which generated two products of slightly different sizes. The DNA ladder and the RT-PCR products were from the same gel. (B) (Upper) The number of known splice junctions detected nears saturation. (Lower) The number of unannotated splice junctions does not saturate at this read depth. (C) Splicing diversity is the highest in hESCs and decreases when cells commit to neural differentiation. The top 500 highly expressed genes shown here were clustered by splice junction diversity (k-means clustering, k = 3) (see SI Text for details). The splice junction diversity value was defined as the number of unique splice junctions detected in the composite gene model given all of the mapped splice junction reads; thus the junction diversity values were normalized for the number of annotated splice junctions in the composite gene model and the number of mapped reads per million. Splice junction diversity is independent of transcript abundance for this set of genes.