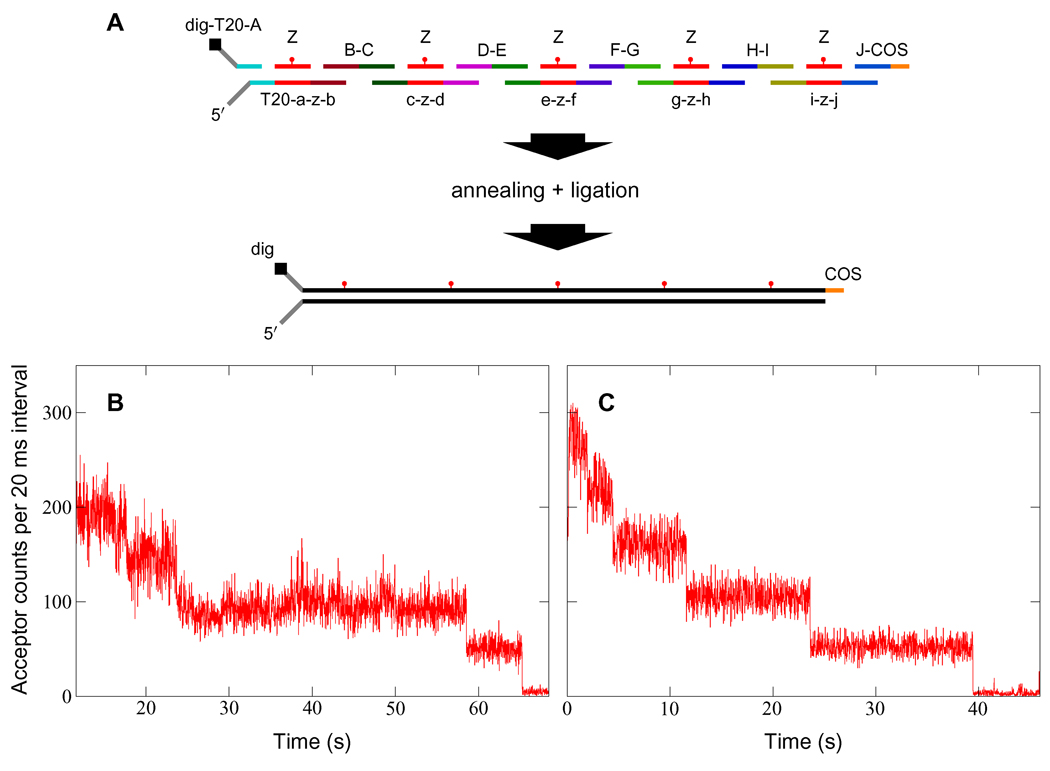
Figure 1.
Synthesis and verification of FRET encoders. (A) DNA self-assembly scheme for 5-period encoders (see Supporting Table 1 for sequences). Each unique sequence is identified with a lower-case letter, and its complement with the corresponding upper-case letter. For example, oligonucleotide E-D (read from 5′ to 3′) pairs only with c-z-d and e-z-f, while e-z-f pairs only with G-F, Z, and E-D. Sequence Z is internally labeled with an acceptor dye. The free 5′ poly(dT) tail facilitates DnaB loading, while the digoxigenin label at the 3′ terminus allows the encoder to be immobilized by attachment to anti-digoxigenin on a fused-silica surface. The 5′ COS overhang is complementary to the 12 bp single-stranded cos site of the λ phage genome. (B and C) Sacrificial photobleaching of four- (B) and five-period (C) encoders by direct excitation at 633 nm verifies complete self-assembly. More than 80% of tested encoders produced the expected photobleaching pattern.