Table 1. Network 2 (E.coli-2): Methods precision for low-to-high recall values.
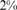
|
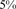
|
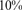
|
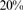
|
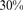
|
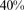
|
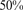
|
|
| mixed-CLR+Inferelator 1.0 |
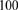
|
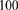
|
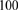
|
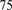
|
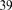
|
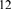
|
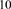
|
| mixed-CLR |
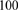
|
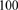
|
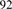
|
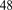
|
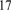
|
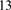
|

|
| dynamic-CLR+Inferelator 1.0 |
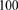
|
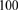
|
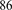
|
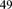
|
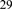
|
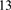
|

|
| dynamic-CLR |
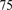
|
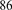
|
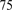
|
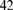
|
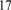
|

|

|
| CLR+Inferelator 1.0 |
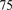
|
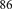
|
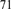
|
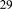
|

|

|

|
| Inferelator 1.0 |
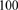
|
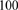
|
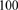
|
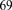
|
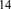
|

|

|
| CLR |
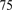
|
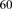
|
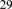
|
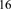
|

|

|

|
In this table we present a more detailed view of performance for our method's best predicted network (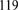 total regulatory interactions with up to
total regulatory interactions with up to  regulators controlling each gene). The table inline method precision [%] at varying degrees of completeness (recall [%]).
regulators controlling each gene). The table inline method precision [%] at varying degrees of completeness (recall [%]).
