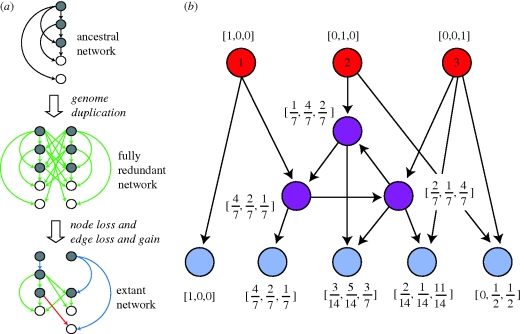
Figure 1.
Regulatory network evolution. (a) Conceptual view of regulatory network evolution after WGD. Immediately after WGD, each ancestral regulatory interaction exists in four copies. As time progresses, both interactions and the TFs and target genes may be lost. Note that the loss of a TF or target also eliminates the interactions that gene possessed. In this thought experiment, we are aware of the ancestral network, meaning that we can distinguish between new interactions (red) and redundant (green) and non-redundant (blue) interactions surviving from WGD. Grey circle, transcription factor; white circle, target gene. In real situations, the ancestral network is unknown, meaning that we cannot make this distinction (adapted from Conant & Wolfe 2008). (b) A simple example of calculating the input diversity of a network. The top-level TFs (red) have minimal input diversity, indicated by vectors with only one non-zero entry. We calculate the vectors for intermediate TFs (purple) and target nodes (blue) iteratively (see §2) until the values converge.