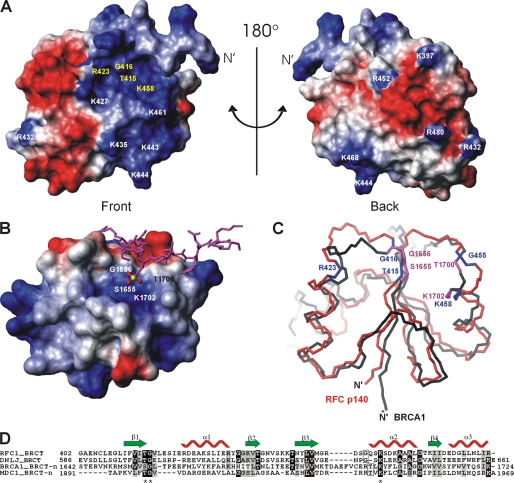
FIGURE 3.
Potential DNA-binding site in p140-(375–480) revealed by amino acid conservation and structure comparison. A, electrostatic potential of the accessible surface of 140(375–480) is shown. Negative potential is colored in red and positive in blue. The absolutely conserved amino acids in the eukaryotic RFC family are in yellow. The orientation of the protein in A is equivalent to Fig. 2D. B, electrostatic surface presentation of the N-terminal BRCT domain (BRCT-n) of BRCA1 (PDB code 1T29) in complex with a phosphoserine peptide (in magenta) colored as in A. The C-terminal BRCT domain is not directly involved in phosphate binding and therefore has been omitted from this figure for clarity. The amino acid residues forming the pocket to accommodate the phosphate moiety (P in yellow and O in red) of phosphoserine are indicated on the surface. C, superposition of p140-(375–480) (red) and the BRCT-n from BRCA1 (black). The orientation of the BRCT-n is identical to that of B. The conserved residues of p140-(375–480) are shown in blue and the phosphate-moiety recognition residues of BRCA1 BRCT-n bound to the phosphoserine peptide are shown in magenta. D, sequence alignment of BRCT domains that bind DNA or phosphopeptides as generated by ClustalW. The sequence alignment was adjusted based on three-dimensional structure alignment using DALILITE (45). The secondary structure of the BRCT domain of p140-(375–480) is depicted. Residues that are >70% identical are shaded black, whereas residues that are >50% similar are shaded gray. The asterisks indicate the three amino acids that bind the phosphate moiety of the bound phosphoserine in the BRCA1 BRCT-n and the MDC1 BRCT-n structures (see the text for the references).