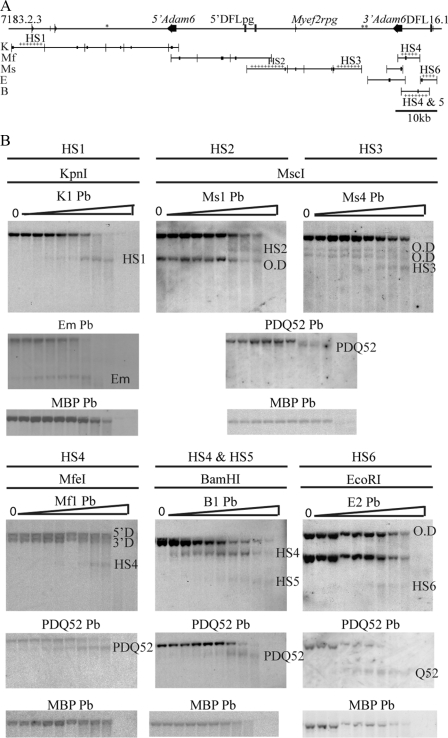
FIGURE 3.
Identification of DNase I-hypersensitive sites in the V-D intergenic region. A, schematic representation of the mouse V-D intergenic region showing restriction fragments and positions of probes used in Southern blots to map DNase I hypersensitivity. Enzymes were as follows: ApaI (A), BamHI (B), BglI (BgI), BglII (BgII), BstEII (BsE), BstXI (BsX), EcoRI (E), KpnI (K), MfeI (Mf), MscI (Ms), PstI (P), PvuII (Pv), SapI (Sa), SpeI (S), XbaI (X), and XcmI (Xc). The positions of the probes (Pb) are indicated by black rectangles on the restriction fragment they were used to detect. The asterisks indicate regions that could not be assayed for DNase I HSs, and crosses indicate restriction fragments in which DNase I HSs were detected. B, Southern blots of DNA from Rag2−/− cell line in which DNase I HS sites were identified. Nuclei were treated with 0–1 unit of DNase/2 × 106 nuclei, indicated by triangles. The HSs are named by their position from the V proximal end of the region. The detection of more than one parental band by the cross-hybridization of the probe to the other duplicon (O.D) in the V-D intergenic region is indicated. Southern blots were reprobed with either the PDQ52 probe or Eμ probe as a positive control, followed by reprobing with the Mbp probe as a negative control.