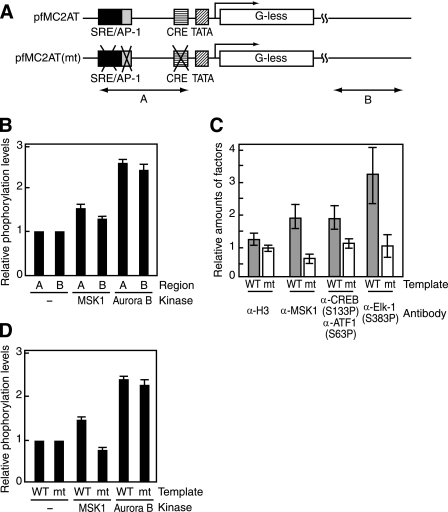
FIGURE 5.
Preferential phosphorylation of histone H3 near the c-fos promoter region. A, wild-type (pfMC2AT) and mutant (pfMC2AT(m)) c-fos templates used for in vitro ChIP assays and the regions amplified by PCR. Region A overlaps with the c-fos promoter, and region B is within the vector pUC19 and ∼1.5 kb downstream from the transcription start site. B, c-fos chromatin was phosphorylated by MSK1 in the presence of activators or by Aurora B and then analyzed by ChIP assays using anti-H3-S10P. The levels of H3-S10P were normalized by the values obtained with anti-histone H3 antibody. The values obtained without any kinase were set to 1. C, in vitro ChIP assays for the occupancies of MSK1 and phosphorylated activators (CREB, ATF1, and Elk-1). Wild-type (WT) and mutant (mt) c-fos templates were assembled into chromatin, and the kinase reactions were done in the presence of MSK1 and the activators. ChIP assays were performed with antibodies against histone H3, MSK1, pCREB(Ser-133) and pElk-1(Ser-383), using the primers for the region A. The anti-pCREB(Ser-133) antibody also recognizes pATF1(Ser-63). Gray and white bars indicate the relative amounts of histone H3, MSK1, and the activators on the wild-type and mutant templates, respectively. D, wild-type and mutant c-fos templates were assembled into chromatin, and the relative levels of H3-S10 phosphorylation in the presence of MSK1 or Aurora B were determined by ChIP assays using anti-H3-S10P and the primers for region A. The error bars represent S.E.