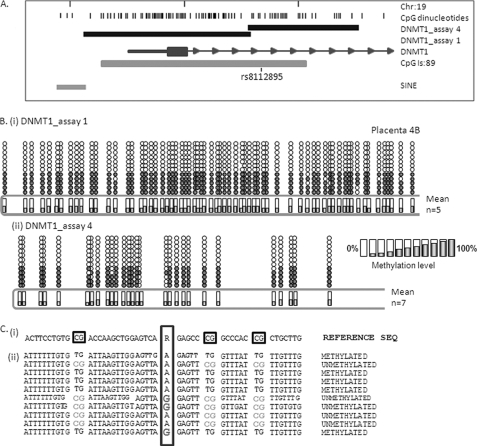
FIGURE 2.
Monoallelic methylation of the human DNMT1 gene. A, location of the DNMT1 CpG island/promoter DNA methylation assays used for bisulfite sequencing. Genomic coordinates on chromosome 19 are shown, along with location of individual CpG dinucleotides (dashes) and CpG island location (gray bar) in relation to the DNMT 1 gene (exon 1 shown as a blue box, intron shown as an arrowed line) according to the University of California Santa Cruz genome browser (hg_18 assembly). The arrows denote transcriptional direction. The location of rs8112895 SNP in the DNMT1_assay_4 is also shown. B, representative DNA methylation of a single full term human placental sample for assay_1 (i) and assay_4 (ii) showing a monoallelic pattern of methylation within the human placenta. Between eight and 12 individual clones were sequenced for each sample. The circles correspond to CpG sites denoted by black dashes in A. Closed circles, methylation; open circles, lack of methylation. Mean methylation levels seen at each CpG site in multiple placental samples for each assay are shown as shaded bars between 0 and 100% (n = 4). Missing circles indicate CpG sites for which no information was obtained. C, DNMT1_4 methylation assay spanning SNP rs8112895 was used for bisulfite sequencing methylation analysis in purified first trimester cytotrophoblast cells. i, reference genomic DNA sequence surrounding rs8112895 SNP (R) and CpG dinucleotides (boxed). ii, individual clone sequences showing the presence of bisulfite-converted CG sites (TG) and unconverted CG sites (CG) in both maternal and paternal alleles from this heterozygous sample. This confirms a lack of parent of origin of the observed DNMT1 promoter methylation.