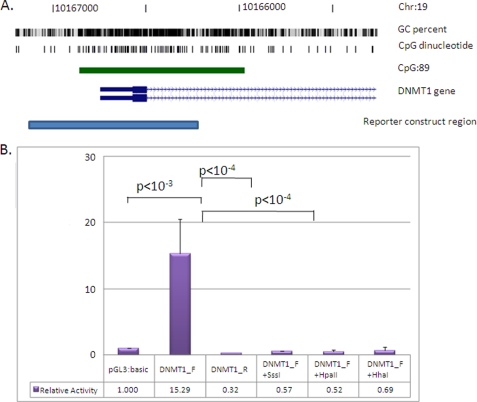
FIGURE 6.
Methylation of the DNMT1 promoter directly attenuates gene transcriptional level. A, position of the cloned DNMT1 promoter region (pale blue bar; −536/+374 relative to the transcription start site). Shown are genome coordinates, GC percentage, location of individual CpG dinucleotides, CpG island (green bar), and DNMT1 gene (exons shown as blue boxes, intron as an arrowed line) according to the University of California Santa Cruz genome browser (hg_18 assembly). The arrows denote transcriptional direction. B, luciferase reporter constructs were transfected into human JAR cells with or without prior in vitro methylation with SssI, HpaII, or HhaI methylases (n = 8 for each). A 15-fold increase in promoter activity was detected for the unmethylated promoter region relative to the promoterless vector. This activity was abolished with in vitro methylation prior to transfection (SssI, HpaII, and HhaI methylases). Luminescence values were normalized to Renilla luciferase to correct for transfection efficiency, and all data are displayed relative to the mean basal promoter activity of the pGL3:basic vector. The error bars denote 95% confidence intervals. Statistical analysis for comparing between transfected groups was carried out using a two-tailed Student's t test with unequal variance.