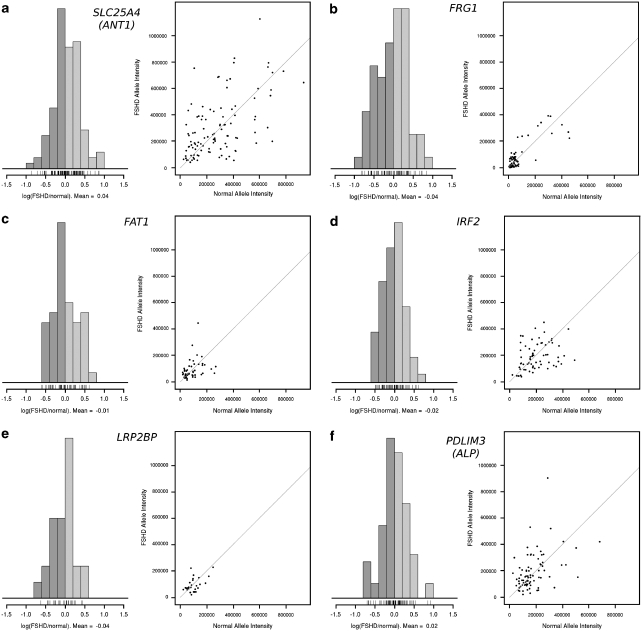
Figure 5.
Ratios of RNA signal intensity between FSHD normal and contracted alleles. Three-color RNA–DNA FISH analysis for the genes (a) SLC25A4 (ANT1), (b) FRG1, (c) FAT1, (d) IRF2, (e) LRP2BP and (f) PDLIM3 (ALP) on FSHD myotube nuclei. Hybridization to D4Z4 was used to identify normal and affected alleles. The intensity of the RNA signal at the FSHD allele was divided by the intensity of the RNA signal at the normal allele and plotted in the histogram on the left. The log of the ratio was used. As such, nuclei with signals of similar intensity (with a ratio near 1) would have a log(ratio) near 0. The greater the fold difference between the two signals was, the greater the distance the log(ratio) would be from 0. The average of the log(ratio) for each gene is listed below the histogram and is very near to 0 for all of the genes. Log(ratios) above 0 (light gray) represent myonuclei in which the affected allele shows greater transcription than the normal allele, whereas ratios below 0 (dark gray) represent myonuclei in which the normal allele shows greater transcription than the affected allele The data depict a balance of transcription between the normal and the affected alleles, and the absence of a cis effect bias to favor either of the alleles. The same procedure was performed using control cell lines with statistically identical results (data not shown). On the scatterplots given on the right, the horizontal axis represents the total probe intensity of the RNA signal at the normal allele. The vertical axis represents the intensity of the signal at the affected allele (with the D4Z4 repeat deletion). Points near the diagonal line represent nuclei in which the two alleles had even signal intensities. Points that stray away from the diagonal line represent nuclei in which one signal was brighter than the other. For all the genes tested, the points remained symmetric across the diagonal line, showing no bias for brighter signals on the affected or unaffected allele.