Table 1.
DNA implementation of the chaotic chemical system due to Willamowsky and Rossle, based on the implementation of Soloveichik et al. (2008). The reaction rates are defined as r1 = 0.03, r2 = r7 = 5 × 104, r3 = r5 = 105, r4 = 0.01, r6 = 0.0165. The implementation uses modules unary0, unary2, binary0, binary1 and binary2, which are defined in a similar fashion to the general modules unaryN and binaryN presented in figure 16. The populations Pg1, … ,Pl7, Pb2, Pb3, Pb, Pb7 and the toehold binding and unbinding rates are chosen to implement accurately the corresponding reaction rates. The populations are passed as parameters to the modules, along with the species A, B, C, where A = (A1,A2,A3), B = (B1,B2,B3) and C = (C1,C2,C3).
| no. | chemistry | DNA molecules |
|---|---|---|
| 1 | 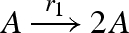 |
unary2(A,Pg1,A,A) |
| 2 | 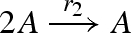 |
binary1(A,A,Pl2,Pb2,A) |
| 3 | 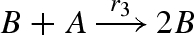 |
binary2(B,A,Pl3,Pb3,B,B) |
| 4 | 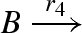 |
unary0(B,Pg4) |
| 5 | 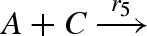 |
binary0(A,C,Pl5,Pb5) |
| 6 | 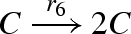 |
unary2(C,Pg6,C,C) |
| 7 | 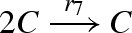 |
binary1(C,C,Pl7,Pb7,C) ) |
