Figure 5.
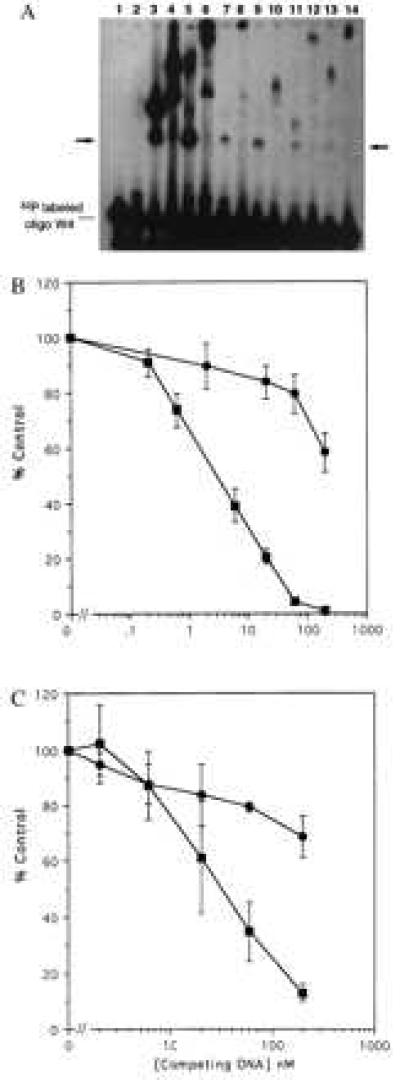
In vitro DNA binding by GST fusion proteins containing library peptides. Candidate DNA binding peptides identified by screening the pXC30 library were fused to GST, expressed, and purified by using standard techniques. (A) Equal amounts (10 μg) of the GST chimeric proteins were incubated with a double-stranded 32P-labeled oligonucleotide (50 fmols) having four WT Sp1 sites. Formation of protein-DNA complexes was monitored by electrophoretic mobility-shift assay. In some cases anti-GST was used to “super-shift” the complexes. The unmarked lane is the free 32P oligonucleotide. The arrow marks the expected migration position of oligonucleotide binding to a fusion protein monomer. Lane 1, unmodified GST protein. Lane 2, two-Zif GST fusion protein. Lane 3, three-Zif fusion protein. Lane 4, the same as lane 3 plus anti-GST antibody. Lane 5, GST fusion with the nonspecific C29 library peptide. Lane 6, the same as lane 5 plus anti-GST. Lanes 7, 9, 11, and 13 are GST fusions with library peptides from clones C4, C7, C23, and C28, respectively. Lanes 8, 10, l2, and 14 are the same as the odd numbered lines plus anti-GST. (B) A radioligand binding competition assay was used to evaluate specificity of DNA binding. Unlabeled double-stranded oligonucleotides having four WT Sp1 sites (▪) or mutated sites (•) were used to competitively displace a 32P oligonucleotide having four WT sites. (Upper) Data for the GST chimera having three Sp1 Zifs. (Lower) Data for the chimera having two Sp1 Zifs linked to the C7 library peptide. The abcissa shows the concentration of unlabeled competitor in nanomolar units. The ordinate shows percent of control radioligand binding (100% = no competitor). The results represent means and standard errors of three determinations.
