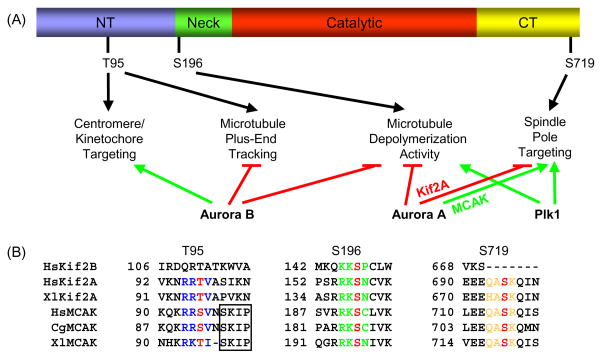
Fig. 2.
Kinesin-13 localization and function is regulated by conserved phosphorylation sites. (A) Schematic of secondary amino acid structure of Kinesin-13s and summary of Aurora B, Aurora A, and Plk1 phosphorylation effects. NT, N-terminal domain; and CT, C-terminal domain. Only the Xenopus MCAK phosphorylation sites are depicted for clarity. Green arrows depict promotion and red lines depict inhibition of localization or activity. (B) The T95, S196, and S719 MCAK phosphorylation sites are highly conserved in Kif2A, whereas only the S196 site is conserved in Kif2B. For HsKif2A, S157 is equivalent to S132 as reported in Knowlton, et al. [70]. Phosphorylation sites are colored red, and the surrounding conserved residues are colored blue for the T95 site, green for the S196 site, and orange for the S719 site. The SxIP microtubule tip localization signal is boxed, and is only conserved in MCAK. Hs, human; Xl, frog (Xenopus laevis); and Cg, Chinese hamster. Genbank accession numbers used for alignment: HsKif2B, NM032559; HsKif2A, NM004520; XlKif2A, BC057698; HsMCAK, NM006845; CgMCAK, U11790; and XlMCAK, BC044976.