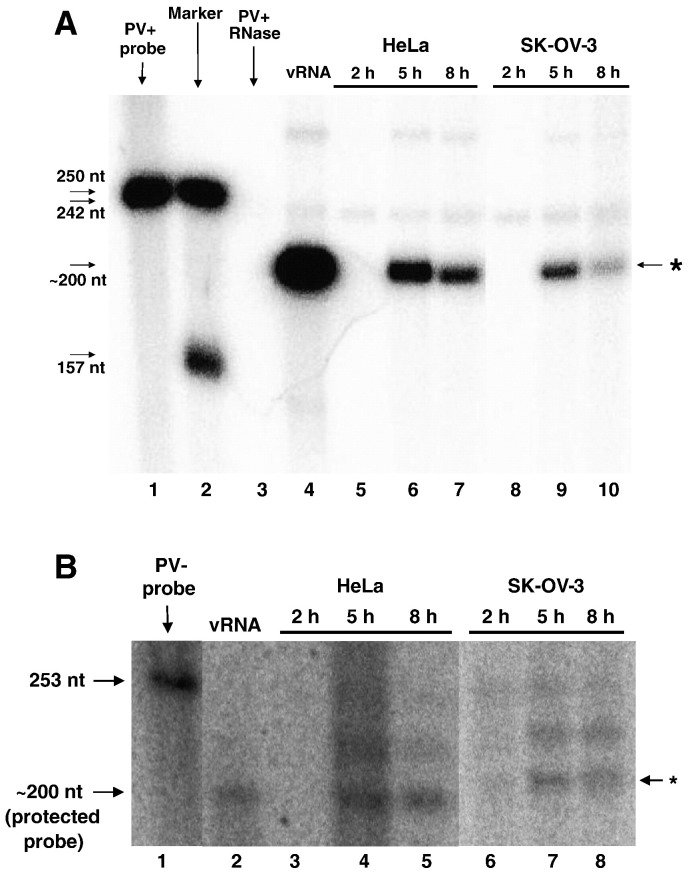
Fig. 3.
Viral RNA accumulation during poliovirus infection of HeLa or SK-OV-3 cell lines. A. Positive-strand viral RNA accumulation was determined using an RNase protection assay as described in Materials and methods. Untreated radiolabeled probe is shown in lane 1. Marker RNAs (with sizes indicated in nucleotides on the left side of the panel) are shown in lane 2. To ensure the RNase used in the assay was active, probe was digested with RNase in the absence of cellular RNA as a control in lane 3. As a positive control for protection of the target virus RNA, 100 ng of purified virion RNA was used for lane 4. Equal amounts (100 ng) of total cellular RNA were protected by 25 fmol of probe complementary to positive-sense poliovirus RNA sequences 5601 to 5809 (lanes 5 to 10). HeLa cells (lane 5 to lane 7) or SK-OV-3 cells (lane 8 to lane 10) were infected with wild type poliovirus. The RNA harvest times (hours post-infection) are indicated at the top of each lane. The protected probe is indicated by the arrow and asterisk on the right side of the panel. These data are representative of three independent experiments. Gel lanes not relevant to this experiment were cropped from this figure, resulting in a spliced image (between lanes 7 and 8). B. Negative-strand viral RNA accumulation was determined using an RNase protection assay as described in Materials and methods. Untreated radiolabeled probe is shown in lane 1. As a positive control for protection of the target virus RNA, 100 ng of purified pT7NPV1 negative-strand RNA was used for lane 2. Equal amounts (1.3 µg) of total cellular RNA were protected by 75 fmol of probe corresponding to positive-sense poliovirus RNA sequences 5601 to 5809 (lanes 3 to 8). HeLa cells (lane 3 to lane 5) or SK-OV-3 cells (lane 6 to lane 8) were infected with wild type poliovirus. The RNA harvest times (hours post-infection) are indicated at the top of each lane. The protected probe is indicated by the arrow and asterisk on the right side of the panel. These data are representative of three independent experiments. Gel lanes not relevant to this experiment were cropped from this figure, resulting in a spliced image (between lanes 1 and 2, and lanes 5 and 6).