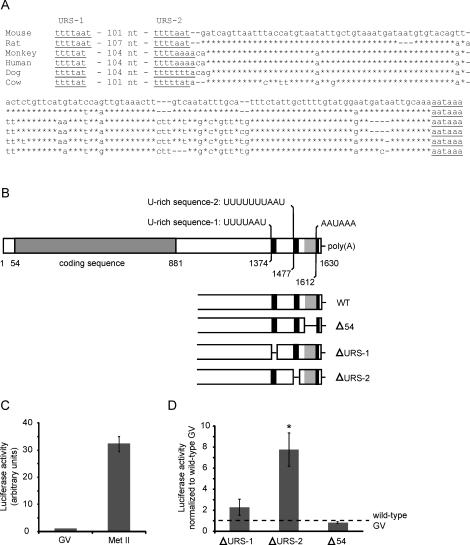
FIG. 3.
Identification and analysis of putative CPE sequences in the 3′-UTR of Slbp mRNA. A) The 3′-UTR of Slbp mRNA in different mammals. Sequences are from Mus musculus (Genbank NM_009193), Rattus norvegicus (XM_001060131), Pan troglodytes (NM_001144566), Homo sapiens (NM_006527), Canis lupus familiaris (XM_545928), and Bos taurus (NM_001045880). The position of first nucleotide and the sequence of URS-1 are shown, followed by the number of nucleotides that separate it from URS-2. The sequence between URS-2 and the polyadenylation signal is shown. Gaps have been introduced to maximize alignment. Nucleotides identical to the mouse are indicated by asterisks. B) The sequence of Slbp mRNA is shown together with the sequence and location of the two URSs identified as putative CPEs and the polyadenylation signal. The lower part of the panel shows the regions that were deleted to generate each mutant 3′-UTR. C) mRNA encoding luciferase linked to the wild-type 3′-UTR of SLBP RNA was synthesized and provided with a short poly(A) tail in vitro and then injected into immature oocytes. Injected oocytes were maintained at the immature (germinal vesicle [GV]) stage, or allowed to mature to metaphase II (Met II). Following a 16-h incubation, luciferase activity was measured; each group contained 15–25 oocytes. The experiment was performed three times. D) As in C, except that URS-1, URS-2, or the 54-nt sequence were deleted from the 3′-UTR. Injected oocytes were maintained at the GV stage, and activity was expressed relative to that in control oocytes injected with the wild-type construct and maintained at the GV stage in the same experiment. Asterisk indicates that the mean differed significantly from the control (t-test, P < 0.05). Error bars indicate the standard error of the mean.