FIGURE 5.
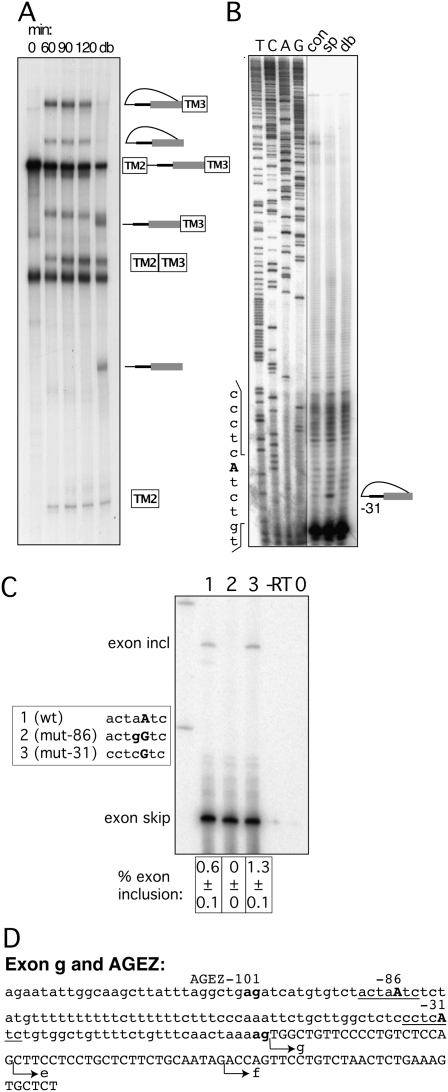
Mapping BPs of exon g. (A) In vitro splicing of construct containing the HTR4 exon g AGEZ. Splicing reactions were incubated from 0 to 120 min. A sample from the 120-min time point was subsequently debranched (lane db). Over the 120-min time course splice products and a single population of lariat and lariat intermediate accumulate. (B) Primer extensions map BP to position −31. Sequences surrounding the BP are depicted on the left side of the gel. Sequencing panel obtained from the same gel but with a different exposure. (C) Mutagenesis of BP in exon trapping construct: lane 1 shows WT splicing pattern with mainly exon skipping and a small amount of exon inclusion. Lane 2—upon disruption of dBP −86 exon inclusion is completely abolished. Lane 3—upon mutation of BP −31 no reduction in exon inclusion is detected. Sequence changes in bold in box beside the gel. Percentage of exon inclusion (n = 3) ± standard deviation is shown underneath the gel. Control lanes are reverse transription reactions without enzyme (−RT) and PCRs without a template (0). (D) Sequence of exon g and its AGEZ: intron sequence in lower case and exon sequence in upper case; first upstream AG dinucleotide and 3′ ss AG in bold. Positions of BPs underlined and BP-As in bold upper case. Alternative 3′ ss of isoforms e and f are indicated by arrows.