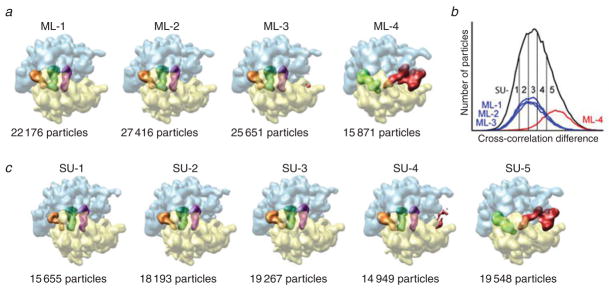
Fig. 7.
Classification of ribosome complexes with or without EF-G by two methods: supervised and unsupervised classification. The experimental data set contains two states of the ribosome, distinguished as follows: (one) contains no EF-G; the A and P sites are occupied by tRNAs in the classic state; and the ribosome is in macrostate I (i.e., unratcheted), the other has EF-G bound; there is a single tRNA in P/E position; and the ribosome is in macrostate II (i.e., ratcheted). (a) Results of classification with the maximum likelihood (ML) method, with four classes specified. (b) Cross-reference between classes from ML and those derived from dividing the correlation histogram obtained by supervised classification. (c) Results of supervised classification using two reference maps, in ribosome macrostates I and II but without EF-G. Data reproduced from Scheres et al. (2007).