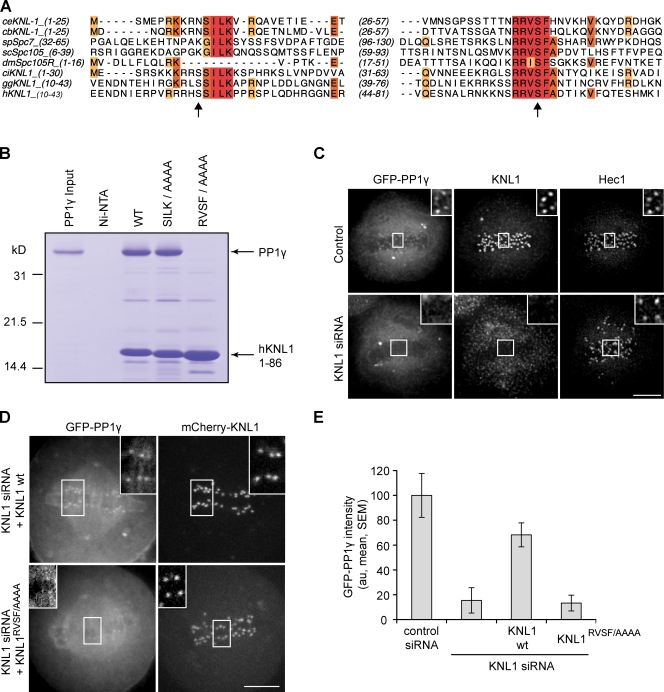
Figure 1.
KNL1 recruits PP1γ to kinetochores. (A) Sequence alignment of C. elegans (ce), Caenorhabditis briggsae (cb), Schizosaccharomyces pombe (sp), Saccharomyces cerevisiae (sc), D. melanogaster (dm), Ciona intestinalis (ci), Gallus galls (gg), and human (h) KNL1 homologues showing conservation of N-terminal [S/G]ILK and RVSF motifs. Arrows indicate Aurora B phosphorylation sites (Welburn et al., 2010). Colors indicate conserved amino acids. (B) Binding of KNL1 to PP1γ depends on the RVSF motif. A Coomassie-stained gel shows binding of PP1γ to Ni-NTA agarose resin alone, or resin bound to either His-tagged hKNL11–86 wild type or mutants for the conserved PP1-binding motifs. (C) HeLa cells stably expressing GFPLAP-PP1γ were fixed and stained for KNL1 and Hec1. PP1γ fails to localize to kinetochores after depletion of KNL1 by siRNA. (D) HeLa cells were transfected with KNL1 siRNA and either siRNA-resistant wild-type (wt) KNL1 or the KNL1RVSF/AAAA mutant, together with GFP-PP1γ. The KNL1RVSF/AAAA mutant fails to restore kinetochore localization of PP1. (E) For cells treated as in D, the intensity of GFP-PP1γ at kinetochores was calculated relative to control cells for each of the conditions indicated. Each bar represents a mean (± SEM) over multiple cells (n ≥ 5), with ≥40 kinetochores analyzed per cell. Images in C and D are maximum intensity projections of confocal stacks; insets show enlarged views (indicated by the boxed regions) of optical sections showing individual kinetochores. Intensity scaling is consistent between all insets, but insets are scaled differently from the original images to show individual kinetochores more clearly. au, arbitrary units. Bars, 5 µm.